Exploring the Microbiota-Gut-Brain Axis for Mental Disorders with Knowledge Graphs
- DOI
- 10.2991/jaims.d.201208.001How to use a DOI?
- Keywords
- Microbiota-gut-brain axis; Gut microbiota; Neurotransmitter; Mental disorder; Knowledge graph; Biomedical ontology
- Abstract
Gut microbiota has a significant influence on brain-related diseases through the communication routes of the gut-brain axis. Many species of gut microbiota produce a variety of neurotransmitters. In essence, the neurotransmitters are chemicals that influence mood, cognition, and behavior of the host. The relationships between gut microbiota and neurotransmitters has received much attention in medical and biomedical research. However, the integration of the various proposed neurotransmitter signal routes that underpin these relationships has not yet been studied well. To unlock the influence of gut microbiota on mental health via neurotransmitters, the microbiota-gut-brain (MGB) axis, we gather the decentralized results in the existing studies into a structured knowledge base. In this paper, we therefore propose a novel Microbiota Knowledge Graph based on a newly constructed knowledge graph for uncovering the potential associations among gut microbiota, neurotransmitters, and mental disorders which we refer to as MiKG. It includes many interfaces that link to well-known biomedical ontologies, e.g. UMLS, MeSH, KEGG, and SNOMED CT, and is extendable by linking to future ontologies to further exploit the relationships between gut microbiota and neurotransmitters. This paper present MiKG, an effective knowledge graph, that can be used to investigate the MGB axis using the relationships among gut microbiota, neurotransmitters, and mental disorders.
- Copyright
- © 2021 The Authors. Published by Atlantis Press B.V.
- Open Access
- This is an open access article distributed under the CC BY-NC 4.0 license (http://creativecommons.org/licenses/by-nc/4.0/).
1. INTRODUCTION
Mental disorders, i.e., mental diseases, lead to a set of problems for both individuals and society [1]. The common mental disorders are depression, anxiety disorders, eating disorders, sleep disorders, bipolar disorder, sex behavior disorder, and autistic disorder [2]. On one hand, for individuals, mental disorders cause significant distress, even functional impairments in many aspects, e.g., emotion, cognition, and behavior [2]. Mental disorders lead to increased risks of suicidal wish, suicide attempt, and suicide [3], and especially major depressive disorder is the main cause of suicide in the world [4]. It is estimated that up to 10% of the person with depression will commit suicide [2]. On the other hand, for society, mental disorders cause heavy economic burdens worldwide [5]. According to the World Health Organization (WHO), approximately one-fifth of all people encounter mental disorders during their lifespan [2]. The number of people with mental disorders is steadily increasing as reported by the Lancet Commission. It is estimated that the costs caused by mental disorders will reach $16 trillion by 2030 [6]. The investigation on the pathogenesis of mental disorders is crucial to treat the patients.
The pathogenesis of mental disorders explicitly includes the role of gut microbiota in the biochemical signaling communications that take place between the gastrointestinal tract and the central nervous system [7,8]. The MGB axis provides a novel path to exploit the pathogenesis of mental disorders and develop appropriate therapeutic strategies. Gut microbiota influences brain-related diseases by interacting with the enteric nervous system and central nervous system [9,10], and may include multiple routes, such as the vagus nerve, the hypothalamic-pituitary-adrenal (HPA) axis, the immune system, cytokines production by immune system, secretion of the short-chain fatty acids (SCFAs), modulation of the neurotransmitters (Figure 1) [11,12]. Changes in these communication routes may result in mental health problems. Neurotransmitters work as a crucial part to regulate the MGB axis, we therefore focus on the signal and communication paths of neurotransmitters, along with the interaction among neurotransmitters, gut microbiota, and mental disorders.
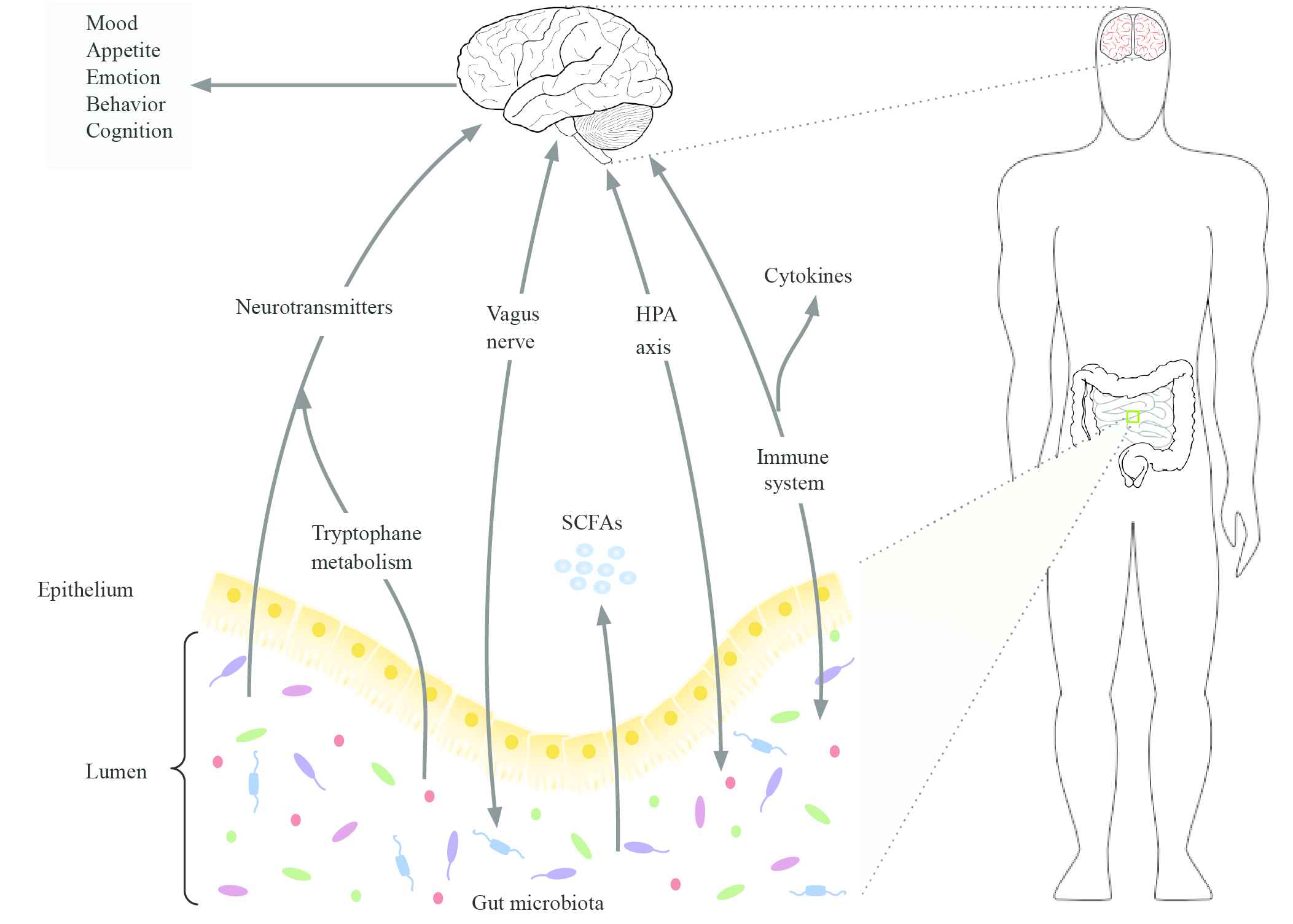
Illustration of the MGB axis. Gut microbiota influence the host health conditions through the bidirectional signal between the gut and the brain. The bidirectional signal communication pathways include the vagus nerve, the HPA axis, the immune system, cytokines production by the immune system, secretion of the SCFAs, modulation of the neurotransmitters.
Gut microbiota affects the level of neurotransmitters of the host, meanwhile, the disruption of neurotransmitters leads to the risk of mental disorders. A potential relationship is that gut microbiota influence the mental health of the host by modulating the level of neurotransmitters in the communication signal of the MGB axis [13]. To investigate this potential relationship, articles on neurotransmitters modulation by gut microbiota have been collected and analyzed [13]. These articles are scattered all over the world and are produced by a multitude of institutions in various formats and standards. Moreover, as these are regular research articles, they are either semi-structured or even unstructured free text. To in-depth exploit the role of gut microbiota and neurotransmitters in the development of mental disorders, we therefore aim to gather the results in the existing studies into a structured knowledge base (i.e., knowledge graphs).
Knowledge graphs are a powerful tool to bring together both structured and unstructured disparate data silos. A knowledge graph is a large-scale semantic network consisting of entities and concepts as well as the semantic relationships among them [14] thereby supporting making better decisions by searching for potential relationships faster. Knowledge graphs have been proven to be useful tools for integrating multiple medical knowledge sources and supporting such tasks as medical decision-making [15], literature retrieval [16], determining medical quality indicators [17], comorbidity analysis [18]. They have been applied to practical problems in bio-medicine, such as learning high-quality knowledge from a knowledge graph based on electronic medical records [19,20], and predicting the relationship between microbes and human diseases [21]. A knowledge graph is sufficient to achieve our aim of gathering the knowledge from a variety of resources and reasoning the potential relationships among gut microbiota, neurotransmitters, and mental disorders [22].
The main contributions of this study are presented as follows:
A new idea to collect the decentralized results of the regulation between neurotransmitters and gut microbiota in existing studies into a structured knowledge base.
A structured way to present the semantic relationships among gut microbiota, neurotransmitters and mental disorders from free text, and makes the knowledge both human-readable and computer-processable.
A graph-based triple store allows the users to carry out semantic query and question answering by using graph-based approaches, and enables the users to obtain more implicit information from the knowledge graph via the ontology reasoning.
A novel knowledge graph model supports researchers to make better research designs by exploring the concealed relations between gut microbiota and neurotransmitters.
2. RELATED WORK
In general, medical ontologies serve as the backbone for the semantic integration. Ontologies work as semantic bridges from primary research to novel therapies. At present, there already exist several medical ontologies, including ontologies for mental diseases [23]. In general, mental disease ontologies do not describe the pathogenesis of the diseases, but only a collection of disease-specific vocabularies and their role relationships. The MGB axis has been proved to be a bidirectional communication between microbiota, the gut, and the central nervous system [9,24]. It has become a potential therapeutic target for many diseases (e.g. mental disorders) [25]. To analyze and understand the complex network of the MGB axis, advanced computer techniques (e.g. knowledge graph and machine learning) are required. Nowadays, psychiatrists and researchers have the opportunity to benefit from complex patterns in brain, behavior, and genes by using these advanced algorithms [26]. Both knowledge graph and machine learning have been applied in various fields of medicine [27]. In this paper, we aim at exploiting the MGB axis for mental disorders by constructing a knowledge graph. We introduce the related work into three parts as follows.
2.1. The MGB Axis in Mental Disorders
Gut microbiota influence brain-related diseases by interacting with the enteric nervous system and central nervous system through multiple communication pathways (Figure 1) [12,28], including the modulation of neurotransmitters. Gut microbiota may influence the mental health of the host by regulating the level of neurotransmitters. On one hand, current studies demonstrate that human gut microbiota are inextricably linked with the mental health of the human host [10]. The composition and diversity of gut microbiota in depressed patients significantly differ from those in healthy controls [29], e.g., chronic administration of suitable probiotics reduced the anxiety- and depression-related behaviors [30,31]. Gut microbiota diversity is involved in the psychopathology of eating [32] and sleeping disorders [33] in humans, while in germ-free fruit flies, walking speed and daily activity are restored by mono-colonization with L. brevis [34]. On the other hand, gut microbiota can produce or modulate the host neurotransmitters directly [25,35]. Lactic acid bacteria such as L. plantarum and L. lactis, form serotonin in vitro [36] and also species of Candida [37], Streptococcus [38], and Enterococcus [39] have the ability to produce serotonin. The family of Bacillus have also been reported to produce dopamine [40] and norepinephrine [41]. Escherichia coli secretes serotonin, norepinephrine, and dopamine in its growth medium [42] while GABA is secreted by certain species of Lactobacillus [13]. Gut microbiota promote the synthesis of histamine [43] and acetylcholine [28] in vivo. The neurotransmitters mentioned above have been implicated most often in the etiological studies of mental disorders [44]. These associations between gut microbiota and mental disorder, on the one hand, and the production of neurotransmitters by various members of the gut microbial community, suggest that manipulation of the gut microbiome may provide a promising path to prevent and treat mental disorders.
2.2. Ontologies for Mental Disorders
Several well-designed biomedical ontologies exist which cover the concepts of mental diseases. Mental Disease Ontology is a repository, which was developed to facilitate representation for all aspects of mental diseases [45]. It also cross-references with other databases like NCI Thesaurus. The NCI Thesaurus is a reference terminology and biomedical ontology that includes the vocabularies of mental diseases related concepts [46]. The Disease Ontology database represents a comprehensive knowledge base of 8043 inherited, developmental, and acquired human diseases [47]. It maps disease and medical vocabularies to several other databases, like MeSH and SNOMED CT [47]. MeSH is the National Library of Medicine's controlled vocabulary thesaurus [48]. SNOMED CT is a systematic collection of medical terms, designed to offer codes, terms, synonyms, and definitions used in clinical documentation and reporting [49]. These databases may be considered to be the most comprehensive collections of medical terms in the world, and they are freely available and updated regularly. They all support users in viewing the role relationships, sibling concepts, and symptoms of the terms. It enables us to obtain extensive information, such as the taxonomy of gut microbiota, by linking with these databases. High-quality ontologies facilitate data aggregation and comparison across different disciplines which may speed up the transformation of primary research into novel therapies [50].
2.3. Knowledge Graph in Mental Disorders
Knowledge graphs allow us to collect and analyze the vast range of medical data available in existing databases, and thereby leverage this knowledge to discover new possible treatment avenues. Knowledge graphs have been proven to be useful tools for integrating multiple medical knowledge sources [22]. Huang et al. proposed a depression-centric knowledge graph which is useful for doctors to explore the relationships among various knowledge resources and to answer realistic clinical queries [51]. Using knowledge graph technology to collect, select, and organize the data, Sang et al. established a recurrent neural network model of known drug therapies, providing an effective way to mining literature for the discovery of new potential drugs, and moreover also providing putative mechanisms of action from literature [52]. Because of the separation of the knowledge base and algorithm programs, it is easy to manage and extend the knowledge base. In this study, a knowledge graph is suitable for us to integrate, analyze, and further extend the knowledge of the MGB axis. It realizes semantic searching, question answering, and visual decision-supporting.
Taken together, constructing a knowledge graph not only gathers existing knowledge resources that enables users to achieve semantic queries and question answering but also supports medical researchers making better decisions to implement novel therapies for mental diseases. The MGB axis is a novel target to investigate the pathogenesis of mental disorders. To exploit the role of the MGB axis in treatment of mental disorders, we therefore aim at constructing a knowledge graph which will enable us to query the associations among gut microbiota, neurotransmitters, and mental disorders. In this paper, we gather available decentralized data from existing databases into a knowledge graph and will highlight its usability by presenting several use-cases.
3. METHODOLOGY
We follow the workflow, as shown in Figure 2, to consolidate data sources into the knowledge graph. First, data source collection. We collect the data sources by doing literature retrieval with a set of keywords. Second, data extraction and structure. We extract the entities and relations from the relevant articles, and structure the relational data in Terse RDF Triple Language (Turtle) format. Third, semantic integration. We describe the method of how we enrich our knowledge base by semantic integrating with other existing ontologies, e.g., UMLS, MeSH, SNOMED CT, and KEGG databases. Finally, constructing a knowledge graph. We input our knowledge base into GraphDB to visualize the knowledge graph and for further analyzing. At this stage, we designed four test cases to demonstrate the performance of our knowledge graph with SPARQL query.
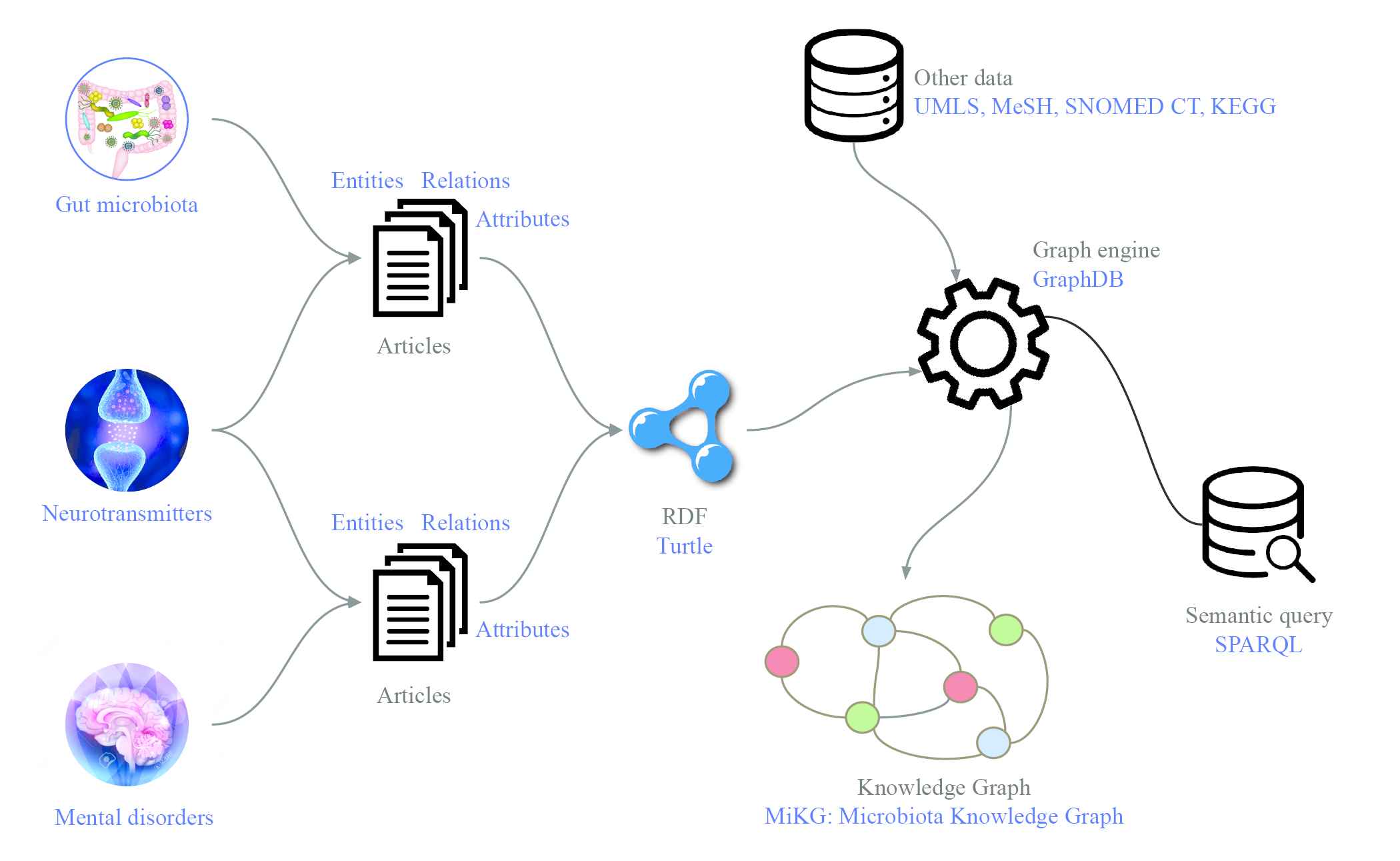
Workflow for constructing the knowledge graph. By doing literature retrieval, we collect articles that studied on the relationship between gut microbiota and neurotransmitters, as well as neurotransmitters and mental disorders. We extract entities, relations and attributes, and structure them into Turtle format. The structured data stored in GraphDB which provides SPARQL query function. The knowledge base enriched by linking with other databases, UMLS, MeSH, SNOMED CT, and KEGG. We named the knowledge graph as MiKG.
3.1. Data Sources Collection
Neurotransmitters play major roles in maintaining human mental health. Their exact numbers are unknown, but more than 200 unique chemical messengers have been identified. In this paper, we take account of six major neurotransmitters: serotonin, dopamine, norepinephrine, GABA, histamine, and acetylcholine, which yield a sufficient data to construct a first knowledge graph of gut microbiota and neurotransmitters. Articles that revealed the relationships between gut microbiota and neurotransmitters, published before December 30, 2019, were identified through a literature search on Google Scholar and PubMed with the keywords: gut microbiota, gut flora, intestinal bacteria, neurotransmitter, serotonin, dopamine, norepinephrine, GABA, histamine, and acetylcholine. With no limitation of study design, all relevant articles were carefully reviewed by three researchers. In the end, thirty-five articles on the regulation between gut microbiota and neurotransmitters were identified for the further extraction of entities and relations. As shown in Table 1, the evidence level of these articles was ranked from A to E according to their strength of the randomized controlled trial design as we presented in Liu and Huang, 2019 [13]. The references of the relationship between neurotransmitters and mental disorders were identified through a literature search on PubMed with keywords: serotonin, dopamine, norepinephrine, GABA, histamine, acetylcholine, anxiety disorders, depressive disorder, sleep disorders, eating disorders, sex behavior disorder, personality disorder, bipolar disorder, autistic disorder, cognition disorders, and learning disorders. If the number of retrieved references for each possible relationship is more than ten, we limit the references to no more than ten which are sorted by the relevance. Relevance is sorted according to the order and frequency of keywords in the title and abstract.
| Level | Design of Study |
|---|---|
| A | Evidence obtained from a systematic review or at least one randomized controlled trials |
| B | Evidence obtained from well-designed pseudo-randomized controlled trials of appropriate size |
| C | Evidence from well-designed trials without randomization, single group pre–post, cohort, time-series studies |
| D | Evidence obtained from case series or nonexperimental studies from more than one center or research group |
| E | Opinions of authorities, based on clinical experience, descriptive studies or reports of expert committees |
Hierarchy of evidence based on the strength of RCTs design [35].
3.2. Data Extraction and Structure
For constructing a knowledge graph, the most important process is to extract the entities and relations from the available data sources. In this paper, the first step, therefore, is to identify and extract entities from the content of relevant articles. Many text mining tools exist that allow users to process structured and unstructured data automatically, however, they all require a large training dataset, and even so will typically fail when encountering new terminology. It was therefore decided to manually obtain highly accurate annotation of entities and semantic relations from the free text which was done by three of us (T.L., X.P., and X.W.). Here, we have several classes of annotations, divided among “entities” (neurotransmitter, mental disorder, gut microbiota, and KEGG pathway), and “relations” (statement, relationship and reference). The six neurotransmitters we mentioned earlier make up the class “Neurotransmitter.” The class “Mental disorders” contains ten common mental disorders: anxiety disorders, depressive disorder, sleep disorders, eating disorders, sex behavior disorder, personality disorder, bipolar disorder, autistic disorder, cognition disorders, and learning disorders. The forty-five gut microbiota entities extracted from articles constitute the class “Gut microbiota.” The collection of the metabolic pathways of the six neurotransmitters is named as “KEGG pathway.” The class “Statement” is used to describe the semantic relational properties between gut microbiota and neurotransmitters, e.g., gut microbiota modulate the level of neurotransmitters. The “Relationship” is used to depict the relations between neurotransmitters and mental disorders, e.g., dopamine is associated with depressive disorder. These extracted entities, concepts, and properties are integrated into one knowledge base in XML format. To make it easier for querying the RDF, we convert the XML data into Terse RDF Triple Language (Turtle) format (Figure 2). Therefore, the resulting set is represented in Turtle syntax, which expresses knowledge in the form of fact statements containing a triplet of entities
3.3. Semantic Integration
We enriched the semantic database by integrating with external biomedical ontologies which are the Unified Medical Language System (UMLS), the Kyoto Encyclopedia of Genes and Genomes (KEGG), the Medical Subject Headings (Mesh), and the Systematized Nomenclature of Medicine—Clinical Terms (SNOMED CT) as shown in Figure 2.
3.3.1. UMLS
The UMLS is a repository of biomedical vocabularies and covers well-known medical terminologies [54]. It integrates over 2 million names for some 900,000 concepts from more than 60 families of biomedical vocabularies, as well as 12 million relations among these concepts. By providing a mapping structure among these vocabularies, it thus allows one to translate among the various terminology systems. Each Metathesaurus concept has a single concept unique identifier (CUI) which link concept data across files. For example, dopamine has a unique CUI number C0013030 that contains its definition, properties, synonym details, and role relationships. In this paper, as shown in Figure 2, CUI is used for characterizing and linking the concepts of gut microbiota, neurotransmitters, and mental disorders with the UMLS.
3.3.2. KEGG
KEGG databases contain multiple databases like pathway database and compound database. The KEGG pathway database is a collection of manually curated pathway maps representing knowledge on molecular interactions, cellular processes, organismal systems, and human disease [55]. Each entry is identified by the map number, e.g., map04728 for dopaminergic synapse that contains dopaminergic synapse-related disease and the cited references. The KEGG compound database is a collection of small molecules, biopolymers, and other chemical substances that are relevant to biological systems. Each entry is identified by the C number, e.g., C00047 for dopamine that contains its chemical structure and associated relationships, along with various links to other databases. These databases are updated regularly. As the Figure 2 shows, we link the neurotransmitters with the KEGG databases for further research purposes.
3.3.3. MeSH
MeSH is a comprehensive controlled vocabulary thesaurus, used for indexing journal articles and books in the life sciences [56]. Most terms in MeSH accompanied by a short description or definition, links to related descriptors, and a list of entry terms (like synonyms or very similar terms) [57]. Each entry is identified by the MeSH Unique ID, for instance, D004298 for dopamine that contains its synonyms and role relationships in all MeSH categories (i.e., tree structures). In tree structures, each term has its unique tree number that allows us to obtain extensive information, such as the taxonomy of gut microbiota and the categories of neurotransmitters. For example, dopamine with tree number D02.092.211.215.406 is the downstream of biogenic monoamines (tree number D02.092.211.215), while dopamine with tree number D02.092.311.342 is the downstream of catecholamines (tree number D02.092.311). In this paper, as shown in Figure 2, MeSH ID is used for linking the concepts of gut microbiota, neurotransmitters, and mental disorders with the MeSH database.
3.3.4. SNOMED CT
SNOMED CT is a standardized, multilingual vocabulary of clinical terminology. It provides codes, terms, synonyms, and definitions of medical terms that are used in clinical documentation and reporting. It currently contains more than 300,000 medical concepts, and each concept is represented by an individual SNOMED CT Identifier (SCTID). For example, dopamine has individual number 412383006 as its unique SCTID. In this paper, as shown in Figure 2, SCTID is used for linking the concepts of gut microbiota, neurotransmitters, and mental disorders with the SNOMED CT database.
3.4. Constructing Knowledge Graph
3.4.1. Knowledge graph in GraphDB
Graphs provide an incredible ability to model potential relationships between information sources and capture linked information (i.e., entity relationships) that many other data models cannot capture, moreover, they enable users to visualize and analyze the data in an interactive and exploratory fashion. We can visualize the knowledge graph and carry out subsequent analysis and optimization work through various graph database management systems such as GraphDB and Neo4j. In this paper, GraphDB was selected to link text and data in big knowledge graphs with its functions such as inserting and transforming any type of data into RDF format, large-scale metadata management, a wide variety of query languages (e.g. SPARQL and SeRQL), full-text search connectors, visualization, semantic similarity search [58]. This database uses graph structures for semantic queries with nodes, edges, and properties to represent and store data. Once the data is represented in graph format, there are various graph analytic techniques to query multiple relationships between entities in the constructed knowledge graph.
3.4.2. SPARQL queries
In this paper, we use RDF as a directed, labeled graph data format to represent the knowledge graph information in GraphDB. SPARQL is used as a query language of the Semantic Web [59]. It contains capabilities for querying required and optional graph patterns along with their conjunctions and disjunctions. SPARQL query contains triple patterns, much like the data itself, which utilize the relationships to quickly navigate any linked data. Generally a SPARQL query contains three components. The PREFIX at the top defines the list of ontologies we use in the query. The SELECT DISTINCT statement defines the variables we want to select (these can be any node in the RDF dataset). The WHERE clause is used to specify a condition while fetching the data from multiple ontologies. Beside this, FILTER (condition) is a clause which can insert into a SPARQL query to filter the results, e.g., limit the data output based on some criteria. The ORDER BY command is used to sort the result set in ascending or descending. The results of SPARQL queries are displayed in various forms. In this paper, we use tables to present the SPARQL result items in the case of a SELECT query.
4. RESULTS
4.1. Construction of the Knowledge Graph
Data from the published studies of gut microbiota on neurotransmitters were retrieved and integrated to create the microbiota knowledge base. This knowledge base of 2,175 triples is stored in GraphDB together with the UMLS (3,081,799 triples), MeSH (13,756,783), and SNOMED CT (4,291,226 triples) databases, as summarized in Table 2. In total, there are 21,131,983 explicit triples. Among the facts, 10,137,028 new records are inferred from the existing triple knowledge, as shown in Table 2. The expansion rate of the knowledge base is 1.48. The whole triples-oriented knowledge base contains 31,269,011 facts altogether, including both explicit and inferred triples (data not shown).
| MiKG | UMLS | MeSH | SNOMED CT | Explicit | Inferred | Total | Expansion Ratio |
|---|---|---|---|---|---|---|---|
| 2,465 | 3,081,799 | 13,756,783 | 4,291,226 | 21,132,273 | 10,136,725 | 31,268,998 | 1.48 |
Triples in knowledge base—MiKG in GraphDB with UMLS, MeSH, and SNOMED CT datasets.
4.2. Knowledge Graph Visualization
In theory, ontology is a “formal, explicit specification of a shared conceptualization” [60]. It consists of a set of concepts (classes), a set of attributes (data type properties), relationships (object properties), and constraints to abstractly represent a specific event [61]. Ontology visualization is an important step in the process of knowledge graph construction. Visualization provides a clear overview of the hierarchy and connections in this knowledge graph. Figure 3 depicts the visualization of our knowledge graph. A knowledge base can be conceptually represented as a collection of terminologies (TBox) and assertions (ABox) [62]. TBox is used to describe a domain of interest by defining classes and properties as a domain vocabulary as shown in blue in Figure 3. ABox is TBox-compliant statements about individuals belonging to these sets, as shown in the pink part in Figure 3. Nodes in orange labeled with MeSH ID link internal classes with external concepts in MeSH via Mesh ID. Therefore, the constructed knowledge graph is able to integrate side information for semantic enrichment. In Figure 3, the labels of edges illustrate the specific attribute and relationship. Arrows are used to show the direction of the relationship from source to target.
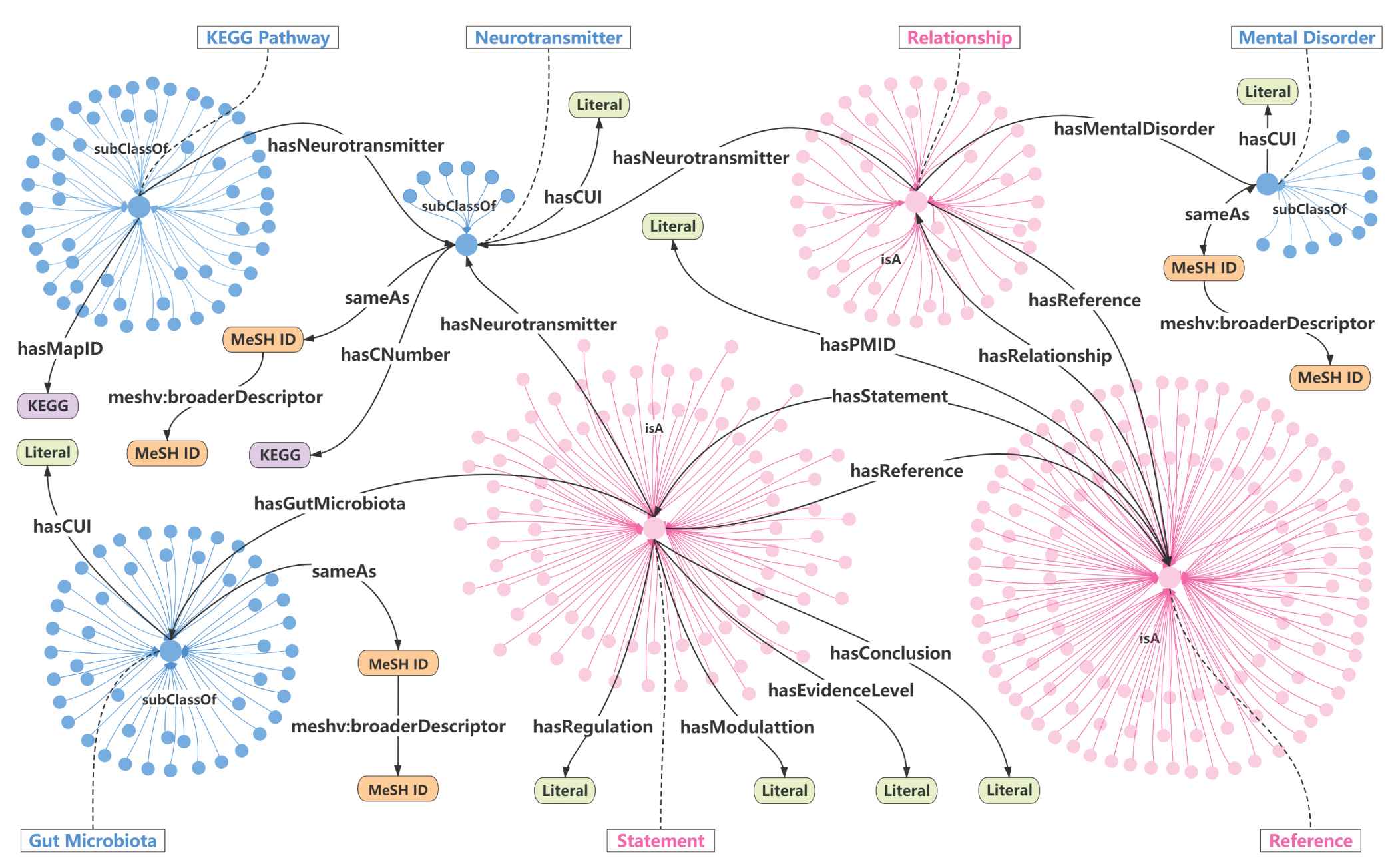
Illustration of the knowledge graph. The blue and pink circular clusters represent TBox (terminologies) and ABox (assertions), respectively. The labels of edges illustrate the specific attribute and relationship. Arrows are used to show the direction of the relationship from source to target. Concepts are linked to UMLS/MeSH through CUI/Mesh ID. Neurotransmitters are linked to KEGG Database through Map-ID and C Number.
4.3. Case Study
To test the proposed knowledge graph for possible associations between gut microbiota, neurotransmitters, and mental disorders, we designed four test cases with various specific conditions. We aim to construct a knowledge graph that is easy to be used by the users, especially those who don't know the knowledge of semantic web standards and the structure of MiKG. Therefore, we designed the SPARQL queries as templates so that users can adjust parameters to suit a particular use case. The MiKG knowledge base, SPARQL query codes, and results of four cases are free available at GitHub.
Listing 1: The SPARQL query code for test case 1-disease-based query. The major depressive disorder is defined as the given condition to return variables.
PREFIX …
SELECT DISTINCT ?GM ?CUI ?Modulation ?NTM ?Ref ?PMID
WHERE {{mikg : Serotonin rdfs:label ?NTM .
?st1 mikg : hasNeurotransmitter mikg : Serotonin ;
mikg : hasGutMicrobiota ?GM ;
mikg : hasModulation ?Modulation ;
mikg : hasReference ?Ref .
?Ref mikg : hasPMID ?PMID .
?GM umls : cui ?CUI}
FILTER EXISTS
{?st2 mikg : hasNeurotransmitter mikg:Dopamine ;
mikg : hasGutMicrobiota ?GM ;
mikg : hasModulation ?Modulation.}
FILTER (lang(?GM) = ‘en’)}
ORDER BY ASC(?GM)
4.3.1. Case 1—disease-based query
In this case, we consider a situation that a person encounters major depressive disorder. We are interested in which species of gut microbiota may cause her depressive disorder by regulating the levels of both serotonin and dopamine. The SPARQL query code is presented in Listing 1, and the query returns 9 results. The obtained results indicate that increase of serotonin and dopamine caused by the eight species of gut microbiota is associated with the development of major depressive disorder of a person. As shown in Table 3, the eight species of gut microbiota namely Bacillus cereus, Burkholderia oklahomensis, Lactobacillus plantarum, Hafnia alvei, Klebsiella pneumoniae, Morganella morganii, Escherichia coli, and Streptococcus thermophilus. Each bacterium has its CUI number in UMLS. These bacteria are involved in the onset of major depressive disorder by increasing the levels of both serotonin and dopamine. This query returns six references that support these facts, as shown in Table 3. Except for two of the references that are not included in PubMed, each of the others has its PMID. Interestingly, there is no gut microbiota species associated with a reduction of serotonin and dopamine in our knowledge graph. In a word, members of the gut microbiota are involved in human's depressive disorder by regulating the levels of both serotonin and dopamine.
| GM | CUI | Modulation | NTM | PMID/Ref |
|---|---|---|---|---|
| Bacillus cereus | C0004590 | Increase | Serotonin; dopamine | 30718848 |
| Burkholderia oklahomensis | C1898603 | Increase | Serotonin; dopamine | 30718848 |
| Escherichia coli | C0014834 | Increase | Serotonin; dopamine | 19845286 |
| Hafnia alvei | C0315259 | Increase | Serotonin; dopamine | Ozogul 2004 |
| Klebsiella pneumoniae | C0001699 | Increase | Serotonin; dopamine | Ozogul 2004 |
| Lactobacillus plantarum | C0317608 | Increase | Serotonin; dopamine | 26522841 |
| Lactobacillus plantarum | C0317608 | Increase | Serotonin; dopamine | Ozogul 2012 |
| Morganella morganii | C0315276 | Increase | Serotonin; dopamine | Ozogul 2004 |
| Streptococcus thermophilus | C0318180 | Increase | Serotonin; dopamine | 23265537 |
Results of test case 1—Disease-based query. The resulting eight species of gut microbiota are associated with development of major depressive disorder in a person. The “CUI” represents the gut microbiota species in UMLS. GM, gut microbiota; NTM, neurotransmitter. N/A for PMID means the reference is not included in PubMed.
4.3.2. Case 2—gut microbiota-based query
Since neurotransmitters and gut microbiota usually affect each other, we therefore consider the situation that neurotransmitters influence the growth of gut microbiota in this test case [13,25]. That is, we aim to study what is the influence of serotonin and norepinephrine on the growth of gut microbiota. We designed the SPARQL query code as shown in Listing 2, and we obtained 9 results by doing the query. As shown in Table 4, serotonin inhibits the growth of Candida albicans and promotes the growth of Enterococcus faecalis, Pichia guilliermondii, Rhodospirillum rubrum, and Saccharomyces cerevisiae. Four different references provide these study results, as presented in Table 4. Two references support that norepinephrine stimulates the growth of Yersinia enterocolitica, Salmonella enterica, Escherichia coli, and Pseudomonas aeruginosa (Table 4). Each bacterium has its CUI number in UMLS and each reference has its PMID in PubMed. Taken together, the growth of gut microbiota is regulated by various neurotransmitters.
| NTM | Regulation | GM | CUI | PMID | Level |
|---|---|---|---|---|---|
| Serotonin | Inhibit | Candida albicans | C0006837 | 16157477 | C |
| Serotonin | Promote | Enterococcus faecalis | C0038404 | 8505913 | C |
| Serotonin | Promote | Pichia guilliermondii | C0319552 | 8505913 | C |
| Serotonin | Promote | Rhodospirillum rubrum | C0035503 | 9702725 | C |
| Serotonin | Promote | Saccharomyces cerevisiae | C0036025 | 21261078 | C |
| Norepinephrine | Promote | Yersinia enterocolitica | C0043406 | 17229058 | C |
| Norepinephrine | Promote | Salmonella enterica | C0445750 | 17229058 | C |
| Norepinephrine | Promote | Escherichia coli | C0014834 | 17229058 | C |
| Norepinephrine | Promote | Pseudomonas aeruginosa | C0033809 | 19517106 | C |
Output records for the test case 2 query. Neurotransmitters influence the growth of gut microbiota. The “CUI” represents the gut microbiota species in UMLS. NTM, neurotransmitter; GM, gut microbiota.
Listing 2: The SPARQL query code of test case 2-gut microbiota based query. We define serotonin and norepinephrine as the specified conditions to return a set of distinct values.
PREFIX …
SELECT DISTINCT ?NTM ?Regulation ?GM ?CUI ?PMID ?Level
WHERE {{mikg : Serotonin rdfs : label ?NTM .
?st1 mikg : hasNeurotransmitter mikg : Serotonin ;
mikg : hasGutMicrobiota ?GM ;
mikg : hasRegulation ?Regulation ;
mikg : hasEvidenceLevel ?Level ;
mikg : hasReference ?Ref .
?GM umls : cui ?CUI .
?Ref mikg : hasPMID ?PMID. }
UNION
{mikg : Norepinephrine rdfs:label ?NTM .
?st2 mikg : hasNeurotransmitter mikg:Norepinephrine ;
mikg : hasGutMicrobiota ?GM ;
mikg : hasRegulation ?Regulation ;
mikg : hasEvidenceLevel ?Level 1;
mikg : hasReference ?Ref .
?GM umls : cui ?CUI .
?Ref mikg : hasPMID ?PMID . }
FILTER (lang(?NTM) =‘en’)}
4.3.3. Case 3—multiple disease-based query
We consider a male patient with three types of disorders: cognition disorders, personality disorder, and learning disorder. To identify which neurotransmitters and gut microbiota are associated with the male patient's mental problems, we design the code to query the association within the knowledge graph, as shown in Listing 3. The SPARQL query generates fifteen results, as shown in Table 5. Results show that fifteen species of gut microbiota relate to. In our knowledge base, serotonin is the only neurotransmitter that is associated with cognition disorders, personality disorder, and learning disorder at the same time. As listed in Table 5, the fifteen gut microbiota species are Paenibacillus, Bacillus cereus, Burkholderia oklahomensis, Acinetobacter baumannii, Lactobacillus plantarum, Bacteroides uniformis, Clostridium ramosum, Hafnia alvei, Klebsiella pneumoniae, Morganella morganii, Escherichia coli, Candida albicans, Lactococcus lactis subsp lactis, Lactococcus lactis subsp cremoris, and Streptococcus thermophilus.
| GM | CUI | Modulation | NTM | PMID/Ref | Level |
|---|---|---|---|---|---|
| Bacillus cereus | C0004590 | Increase | Serotonin | 30718848 | B |
| Burkholderia oklahomensis | C1898603 | Increase | Serotonin | 30718848 | B |
| Acinetobacter baumannii | C0314787 | Increase | Serotonin | 30718848 | B |
| Paenibacillus | C1011299 | Increase | Serotonin | 30718848 | B |
| Lactobacillus plantarum | C0317608 | Increase | Serotonin | 26522841 | B |
| Bacteroides uniformis | C0314925 | Increase | Serotonin | 25860609 | B |
| Clostridium ramosum | C0315111 | Increase | Serotonin | 30718836 | B |
| Escherichia coli | C0014834 | Increase | Serotonin | 19845286 | C |
| Candida albicans | C0006837 | Increase | Serotonin | 16157477 | C |
| Lactococcus lactis subsp lactis | C1449851 | Increase | Serotonin | Ozogul 2012 | C |
| Lactococcus lactis subsp cremoris | C0544170 | Increase | Serotonin | Ozogul 2012 | C |
| Streptococcus thermophilus | C0318180 | Increase | Serotonin | 23265537 | C |
| Hafnia alvei | C0315259 | Increase | Serotonin | Ozogul 2004 | C |
| Klebsiella pneumoniae | C0001699 | Increase | Serotonin | Ozogul 2004 | C |
| Morganella morganii | C0315276 | Increase | Serotonin | Ozogul 2004 | C |
Output records for the test case 3 query. By regulating the level of serotonin, fifteen species of gut microbiota are associated with cognition disorders, personality disorder, and learning disorder. GM, gut microbiota; NTM, neurotransmitter; MD, mental disorder.
Listing 3: The SPARQL query code of test case 3 - neurotransmitter based Query. Three different mental disorders are set as conditions to call a list of variables.
PREFIX …
SELECT DISTINCT ?GM ?CUI ?Modulation ?NTM ?PMID ?Level
WHERE {mikg : Cognition-disorders rdfs : label ?MD1 .
mikg : Personality-disorder rdfs : label ?MD2 .
mikg : Learning-disorder rdfs : label ?MD3 .
{?re1 mikg : hasMentalDisorder mikg : Cognition-disorders ;
mikg : hasNeurotransmitter ?NTM ;
?st mikg : hasNeurotransmitter ?NTM ;
mikg : hasGutMicrobiota ?GM ;
mikg : hasEvidenceLevel ?Level ;
mikg : hasModulation ?Modulation ;
mikg : hasReference ?Ref .
?Ref mikg : hasPMID ?PMID .
?GM umls : cui ?CUI . }
FILTER EXISTS
{?re2 mikg : hasMentalDisorder mikg : Personality-disorder ;
mikg : hasNeurotransmitter ?NTM . }
FILTER EXISTS
{?re3 mikg : hasMentalDisorder mikg : Learning-disorder ;
mikg : hasNeurotransmitter ?NTM . }
FILTER (lang(?MD1) =‘en’)
FILTER (lang(?MD2) =‘en’)
FILTER (lang(?MD3) =‘en’)}
4.3.4. Case 4—neurotransmitter-based query
Histamine is a typical neurotransmitter that relates to mental disorders. Histamine levels are also closely tied to the composition and diversity of the gut microbiota [63]. On the one hand, a wide diversity of bacteria from the human gut produces and degrades histamine [64]. On the other hand, gut microbiota promote the human basophil leukocytes cells and mast cells to release histamine that influence the host immunological processes [65]. In this test case, we therefore aim to investigate the association between gut microbiota and mental disorders when histamine is increased or reduced. We design the SPARQL code to query this association in our knowledge graph, as shown in Listing 4. The association between gut microbiota and histamine are ranked to 5 evidence levels from A to E (cf. Subsection 3.1), we only consider the association of evidence level C for specific in this test case. As shown in Table 6, histamine is regulated by three species of gut microbiota, i.e., Escherichia coli, Lactobacillus vaginalis, and Streptococcus thermophilus. By regulating histamine levels, these three bacteria are involved in the development of five mental disorders which are anxiety disorders, bipolar disorder, depressive disorder, learning disorders, and sleep disorders.
| GM | Modulation | NTM | PMID | Level | MD | |
|---|---|---|---|---|---|---|
| 1 | Escherichia coli | Increase | Histamine | 2412960 | C | Anxiety disorder |
| 2 | Escherichia coli | Increase | Histamine | 2412960 | C | Bipolar disorder |
| 3 | Escherichia coli | Increase | Histamine | 2412960 | C | Depressive disorder |
| 4 | Escherichia coli | Increase | Histamine | 2412960 | C | Learning disorders |
| 5 | Escherichia coli | Increase | Histamine | 2412960 | C | Sleep disorders |
| 6 | Lactobacillus vaginalis | Increase | Histamine | 26394683 | C | Anxiety disorder |
| 7 | Lactobacillus vaginalis | Increase | Histamine | 26394683 | C | Bipolar disorder |
| 8 | Lactobacillus vaginalis | Increase | Histamine | 26394683 | C | Depressive disorder |
| 9 | Lactobacillus vaginalis | Increase | Histamine | 26394683 | C | Learning disorders |
| 10 | Lactobacillus vaginalis | Increase | Histamine | 26394683 | C | Sleep disorders |
| 11 | Streptococcus thermophilus | Increase | Histamine | 23265537 | C | Anxiety disorder |
| 12 | Streptococcus thermophilus | Increase | Histamine | 23265537 | C | Bipolar disorder |
| 13 | Streptococcus thermophilus | Increase | Histamine | 23265537 | C | Depressive disorder |
| 14 | Streptococcus thermophilus | Increase | Histamine | 23265537 | C | Learning disorders |
| 15 | Streptococcus thermophilus | Increase | Histamine | 23265537 | C | Sleep disorders |
Output records for the test case 4 query. Three gut microbiota species associated with six mental disorders by modulating the concentration of histamine. The “Level” refers to the evidence level of histamine modulated by gut microbiota. GM, gut microbiota; NTM, neurotransmitter; MD, mental disorder.
Listing 4: The SPARQL query code of test case 4 - neurotransmitter based Query. Histamine is defined as the specific condition to retrieve various values.
PREFIX …
SELECT DISTINCT?GM ?Modulation ?NTM ?PMID ?Level ?MD
WHERE {{mikg : Histamine rdfs : label ?NTM .
?re mikg : hasNeurotransmitter mikg:Histamine ;
mikg : hasMentalDisorder ?MD .
?st mikg : hasNeurotransmitter mikg : Histamine ;
mikg : hasGutMicrobiota ?GM ;
mikg : hasModulation ?Modulation ;
mikg : hasEvidenceLevel ?Level ;
mikg : hasReference ?Ref .
?Ref mikg : hasPMID ?PMID .
FILTER (lang(?NTM) =‘en’)
FILTER (?Level =‘C’)}}
ORDER BY ASC(?GM)
5. DISCUSSION AND OUTLOOK
In this study, we constructed a knowledge graph to explore the role of the MGB axis, especially the neurotransmitters pathway, in mental disorders. We extracted the entities of gut microbiota and neurotransmitters, along with their relational properties, from the free text in articles. Due to the sources of knowledge extraction being diverse, the knowledge obtained by different sources is not compatible, which puts forward a demand for knowledge integrating. We linked the gut microbiota with mental disorders via neurotransmitters through the construction of the knowledge graph. The knowledge graph semantic database was enriched by integrating it with other biomedical ontologies, thereby providing users relevant and accurate information and relationships, as a solid basis for exploring the role of the neurotransmitters in the pathogenesis of mental disorders.
The relative success of our knowledge graph attribute to the distinctive features that not presented in other knowledge graphs. Its main feature is to facilitate users to explore the influence of gut microbiota on mental health. So far, there have been many applications of knowledge graphs in the medicine, such as drug discovery [52], targets predication [66], and disease classification [67]. There are few studies on knowledge graphs and mental disorders. Huang et al. crated a depression knowledge graph by integrating various knowledge resources about depression (e.g., clinical trials, antidepressants, medical publications, clinical guidelines, etc.) [51]. It provides a data infrastructure to explore the relationship among various knowledge and data sources about depression. What is lacking is a knowledge graph that can be used to investigate the pathogenesis of mental diseases. Current studies indicate that the MGB axis plays a significant role in maintaining mental health of the host. We therefore construct the knowledge graph MiKG to unlock the influence of gut microbiota on mental health via neurotransmitter, one of the pathways of the MGB axis.
The performance of a knowledge graph supporting the effective discovery of implicit relationships is no doubt important. In a domain knowledge graph database, relationships can be divided into explicit relationships and implicit relationships. Explicit relationships are relationships that can be extracted directly from the original data, and implicit relationships are dynamic relationships that need to be calculated through traversing the knowledge graph. We conduct 4 case studies to demonstrate the discovery potential of our knowledge graph by SPARQL querying. Simply, we can semantically search the explicit relationships, such as the relationship between gut microbiota and neurotransmitters, as we performed in test case 1 and case 2 (Subsubsections 4.3.1 and 4.3.2). For discovering the implicit relationships, we can explore the potential influence of gut microbiota on mental health via neurotransmitters, as we did in test case 3 and case 4 (Subsubsections 4.3.3 and 4.3.4). By enriching the knowledge graph with other existing databases we enable users access to other databases and obtains more implicit relationships. For example, we integrate the KEGG database of pathways into our knowledge base, so that enables the discovery of which metabolic pathways the gut microbes influence are related to mental health. We have the complete entities and concepts of gut microbiota, neurotransmitters, and mental disorders in the knowledge base which extended by linking to UMLS, MeSH, and SNOMED CT. Crucially, each of those databases do not describe any relation between, e.g., gut microbiota and mental disorder; our MiKG aims to fill this gap. Taken together, our knowledge graph has the discovery potential to find implicit relationships as we expected.
Our knowledge graph, however, has limitations. Gut microbiota has an impact on mental health by regulating neurotransmitters, but that is not as simple as one bacterium and one neurotransmitter. As we know, the gut microbiome consists of many thousands of species of microorganisms [68], while dozens of neurotransmitters related to mental health have been discovered so far [69]. Besides, over 300 different mental disorders cataloged in the DSM-5 (Diagnostic and Statistical Manual of Mental Disorders) [70]. However, the number of explicit relations between gut microbiota, neurotransmitters, and mental disorders retrieved from literature current MiKG was limited by the necessity of manual extraction from literature, as this allowed us to focus on a small but accurate sub-set of the data as a proof of principle.
The accuracy and completeness of extracted relations in the MiKG, also limits the precision and reliability of the semantic search results. Therefore, our future work will emphasize improving the accuracy and completeness of the knowledge base. To do this effectively while not sacrificing too much accuracy, we will adopt text-mining approaches and relations extraction tools. Manual processing ensures high accuracy of the data but cannot cover the complete knowledge domain. Taken together, our future work will apply advanced automated processing to discover the relations between gut microbiota, neurotransmitters, and mental disorders, and update the knowledge base automatically.
6. CONCLUSION
To predict the relationships between gut microbiota and mental disorders with neurotransmitters as the linking element, we first constructed a knowledge graph by integrating the disparate knowledge from existing biomedical studies in a semantic way. Such a knowledge graph, MiKG, benefits us to discover implicit knowledge by semantic query and reasoning. We designed various test cases to demonstrate the potential identification and prediction performance of the knowledge graph by using the SPARQL query. Results indicate that MiKG is a powerful tool for uncovering potential associations among gut microbiota, neurotransmitters, and mental disorders. It not only effectively supports identifying explicit relationships, such as relationships between gut microbiota and neurotransmitters, but importantly also enables to infer implicit relationships, such as the influence of gut microbiota on mental health via neurotransmitters. In summary, our novel MiKG knowledge graph developed here, is an effective tool to identify, explore, and predict the relationships among gut microbiota, neurotransmitter, and mental disorders. It has the potential to infer reasonable hypotheses, thereby accelerating the development of new treatment for mental disorders, and benefit the field of the MGB axis investigation.
COMPETING INTERESTS
The authors declare no competing interests.
AVAILABILITY OF DATA AND CODE
The data and code that support the findings of this study are openly available at GitHub.1
AUTHORS' CONTRIBUTIONS
T.L. and Z.H. designed the project. T.L., X.P., and X.W. performed the data sources collection, data extraction and structure. T.L., X.P., and Z.H. designed and performed the test cases. Z.H., K.A.F., and J.H. supervised and directed the work continuously. T.L. wrote the manuscript with contributions from all co-authors. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
REFERENCES
Cite this article
TY - JOUR AU - Ting Liu AU - Xueli Pan AU - Xu Wang AU - K. Anton Feenstra AU - Jaap Heringa AU - Zhisheng Huang PY - 2020 DA - 2020/12/15 TI - Exploring the Microbiota-Gut-Brain Axis for Mental Disorders with Knowledge Graphs JO - Journal of Artificial Intelligence for Medical Sciences SP - 30 EP - 42 VL - 1 IS - 3-4 SN - 2666-1470 UR - https://doi.org/10.2991/jaims.d.201208.001 DO - 10.2991/jaims.d.201208.001 ID - Liu2020 ER -
