TMRGM: A Template-Based Multi-Attention Model for X-Ray Imaging Report Generation
 , Yu Zhang, Zhen Guo
, Yu Zhang, Zhen Guo , Jiao Li*,
, Jiao Li*, 
Xuwen Wang and Yu Zhang are co-first authors.
- DOI
- 10.2991/jaims.d.210428.002How to use a DOI?
- Keywords
- Chest X-ray; Deep learning; Thoracic abnormality recognition; Medical imaging report generation; Attention mechanism; Medical imaging report template
- Abstract
The rapid growth of medical imaging data brings heavy pressure to radiologists for imaging diagnosis and report writing. This paper aims to extract valuable information automatically from medical images to assist doctors in chest X-ray image interpretation. Considering the different linguistic and visual characteristics in reports of different crowds, we proposed a template-based multi-attention report generation model (TMRGM) for the healthy individuals and abnormal ones respectively. In this study, we developed an experimental dataset based on the IU X-ray collection to validate the effectiveness of TMRGM model. Specifically, our method achieves the BLEU-1 of 0.419, the METEOR of 0.183, the ROUGE score of 0.280, and the CIDEr of 0.359, which are comparable with the SOTA models. The experimental results indicate that the proposed TMRGM model is able to simulate the reporting process, and there is still much room for improvement in clinical application.
- Copyright
- © 2021 The Authors. Published by Atlantis Press B.V.
- Open Access
- This is an open access article distributed under the CC BY-NC 4.0 license (http://creativecommons.org/licenses/by-nc/4.0/).
1. INTRODUCTION
Medical imaging data is the key basis for early screening, diagnosis, and treatment of diseases. In a real clinical scenario, professional radiologists review and analyze medical images empirically, then describe imaging findings and write the diagnosis conclusions in semi-structured reports. However, the rapid growth of medical imaging data brings heavy workload to radiologists for image reading and report writing. How to assist doctors in medical image interpretation has become an important and challenging task for computers.
In the last decade, the interdisciplinary research and application of medical imaging and advanced intelligence technology are growing rapidly [1]. Driven by large-scale open access image dataset, deep learning, represented by convolutional neural network (CNN) [2] and recurrent neural network (RNN) [3], push forward the development of computer-aided diagnosis (CAD) systems [4], which can effectively process large-scale multimodal medical images, detect abnormal lesions, and distinguish the nature of the lesion [5–9]. In the computer vision area, deep natural language processing (NLP) technology can be used to describe images by combining the image features with the text features. Inspired by this, more complex cognitive tasks such as visual captioning and medical image report generation have attracted growing attention in recent years [28–30].
However, despite the state-of-the-art progress, it is still challenging to generate clinically readable and interpretable reports. For example, existing methods perform better on generating short descriptions of images, but incapable of diversifying language and depicting long complex structures [10,11]. Linguistically, most studies treat visual words and nonvisual words equally (such as “there,” “evidence,” “seen,” “to,” etc.), while the latter have no correlation with any image features and may be misleading for text generation. Additionally, in the real clinical setting, radiologists often write normal reports based on unified templates, and reports of healthy individuals only describe normal organ or structures. However, most studies treat the reports of healthy individuals and abnormal ones with similar methods. There is little difference between the generated reports of healthy individuals and sick ones, especially underperform on depicting rare abnormal findings.
In addressing this problem, we proposed a novel framework for chest X-ray image interpretation and report generation by exploiting the different structure of healthy/abnormal reports. The major contributions of this paper are summarized as follows. (1) We proposed template-based multi-attention report generation model (TMRGM), a new template-based multi-attention mechanism for chest X-ray report generation, which utilize different strategies to generate imaging reports for healthy individuals and abnormal ones respectively. (2) To generate chest X-ray imaging reports for healthy individuals, we manually constructed a library of chest X-ray report templates. (3) To generate chest X-ray imaging reports for abnormal individuals, we integrate image features and text features via co-attention mechanism and adaptive attention mechanism. The model can automatically choose whether to generate report text based on image features, sentence topics, or text features. (4) We verified the performance of chest lesion recognition and report generation based on the public available IU X-ray dataset (Open I) [18].
2. RELATED WORKS
2.1. Medical Imaging Datasets
In recent years, deep neural networks have shown great potential in challenging tasks of medical image processing [12,13]. The rapid improvement partly depends on the publicly accessible medical imaging datasets that covering multimodal and various body parts with quality annotation. In particular, images concerning chest diseases, e.g., chest X-rays and chest CT scan are commonly used for clinical screening and diagnosis, and account for a large proportion in public datasets.
For instance, the NIH released the ChestX-ray14 dataset for thoracic lesion detection [14]. The National Cancer Institute (NCI) released the LIDC–IDRI dataset for early cancer detection in high-risk populations [15] and Data Science Bowl 2017 [16], the high-resolution CT scan data for lung cancer prediction. The Stanford University present CheXpert [17], a large-scale dataset that contains 224,316 chest radiographs of 65,240 patients. OpenI [18] contains chest X-ray reports of 3,955 patients and 7470 chest X-ray images, which has become the benchmark of the current research on imaging report generation. Recently, MIT released MIMIC-CXR-JPG v2.0.0 [19], a large dataset of 377,110 chest X-rays associated with 227,827 imaging studies sourced from the Beth Israel Deaconess Medical Center. In addition, during the outbreak time of COVID-19, many small-scale datasets are released for developing AI-based diagnosis models of COVID-19. For instance, Yang et al. build an open-sourced dataset COVID-CT [41], which contains 349 COVID-19 CT images from 216 patients and 463 non-COVID-19 CT. Li et al. introduced COV-CTR [42], a COVID-19 CT report dataset which contains 728 images collected from published papers and their corresponding paired Chinese reports.
2.2. Thoracic Lesion Recognition
In the early stage of image recognition, some feature extraction methods, such as histogram of oriented gradients (HOG) and scale invariant feature transform (SIFT) were mainly used to classify and recognize the extracted features through classifiers [43]. Early image recognition tasks are targeted at specific recognition objects, without generalization ability, and the sample size is small, so it is difficult to meet high recognition requirements in practical application.
Thoracic Lesion Recognition (TLR) has long been a research focus in CAD. According to the types of identified lesions, TLR methods can be divided into two categories. One is single thoracic lesion recognition (sTLR), which focuses on the imaging characteristics of a particular type of lesion. It can assist the early screening and diagnosis of a specific disease, e.g., the pulmonary nodule detection [20,21]. The other one is multiple thoracic lesion recognition (mTLR), which target multiple types of disease or lesion, such as pulmonary nodules, pneumonia, pneumothorax, pleural effusion, atelectasis, pulmonary abscess, pulmonary tuberculosis, etc. The mTLR is more consistent with the radiologists' way of reading images, and can better support comprehensive diagnosis.
There are commonly two steps in mTLR: (1) multi-label classification (MLC) of thoracic lesions revealed in chest radiography; (2) thoracic lesion localization, which identifies specific regions and profile of abnormal lesions in chest radiography. In recent years, deep learning models start to outperform conventional statistical learning approaches [43,44] in the TLR task. A representative work is the CheXNet developed by Ng et al. [22], a 121-layer dense convolutional neural network (dense CNN), which detect 14 chest diseases simultaneously based on the ChestX-Ray14 data set. Bar et al. [23] used the pretrained CNN model to extract the high-dimensional features of medical images, and combined them with general GIST feature and bag-of-visual words (BoVW) features as the input of support vector machine (SVM) to detect thoracic lesions. Wang et al. [14] developed a Ddep convolutional neural network (DCNN) for mTLR. Yao et al. [24] constructed a DenseNet-long short-term memory (DENsenet-LSTM) model to identify the 14 thoracic lesions by utilizing latent correlation between different lesions in chest X-ray images.
2.3. Visual Captioning and Medical Image Report Generation
Visual captioning aims at generating a descriptive sentence for a given image or video. Most state-of-the-art methods generated sequences based on the CNN-RNN architectures and attention mechanisms [45–47]. In addition to the one-sequence generation in early studies, some efforts have been made for generating longer paragraphs [11], which inspires the research of medical image report generation. However, medical image reports are more professional and informative than natural image captions, which poses greater challenge on generating clinically readable reports. Shin et al. first proposed a variant of CNN-RNN framework to predict lesion tags of chest X-ray images [25]. Wang et al. [26] developed Latent Dirichlet Allocation-based topic models for imaging report generation. Kisilev et al. [27] proposed a CNN-based method for generating reports of classified mammography images. Wang et al. proposed the TieNet model [28], integrating the multi-attention model into the end-to-end CNN-RNN framework for performing disease classification and generating simple imaging reports. Jing et al. [29] constructed a hierarchical language model equipped with co-attention to better model the paragraphs, but it tend to produce normal findings. They went further to explore the complex structures of reports, and proposed a two-stage strategy that models the relationship between Findings and impression [48]. Li et al. [30] proposed KERP, a knowledge-driven imaging report generation model, which constructed a graph transformer (GTR) for the dynamic transformation of text features and image features.
The difference between our proposed model and existing methods lies in that we classified chest X-rays into healthy or abnormal individuals based on MLC module, then we combined report templates with multi-attention-based hierarchical LSTM model and generate reports respectively according to the nature of the given image (healthy/abnormal). In addressing the problem that the nonvisual feature words are difficult to align with the image features, TMRGM- generated visual words and nonvisual words separately based on features from different modality.
3. METHOD
As shown in Figure 1, the proposed framework is comprised of three modules: (1) the chest X-ray classification (healthy/abnormal) module based on multi-label thoracic lesion recognition; (2) template-based report generation module for healthy individuals; and (3) multi-attention-based Hierarchical LSTM module for abnormal individuals [32–34].
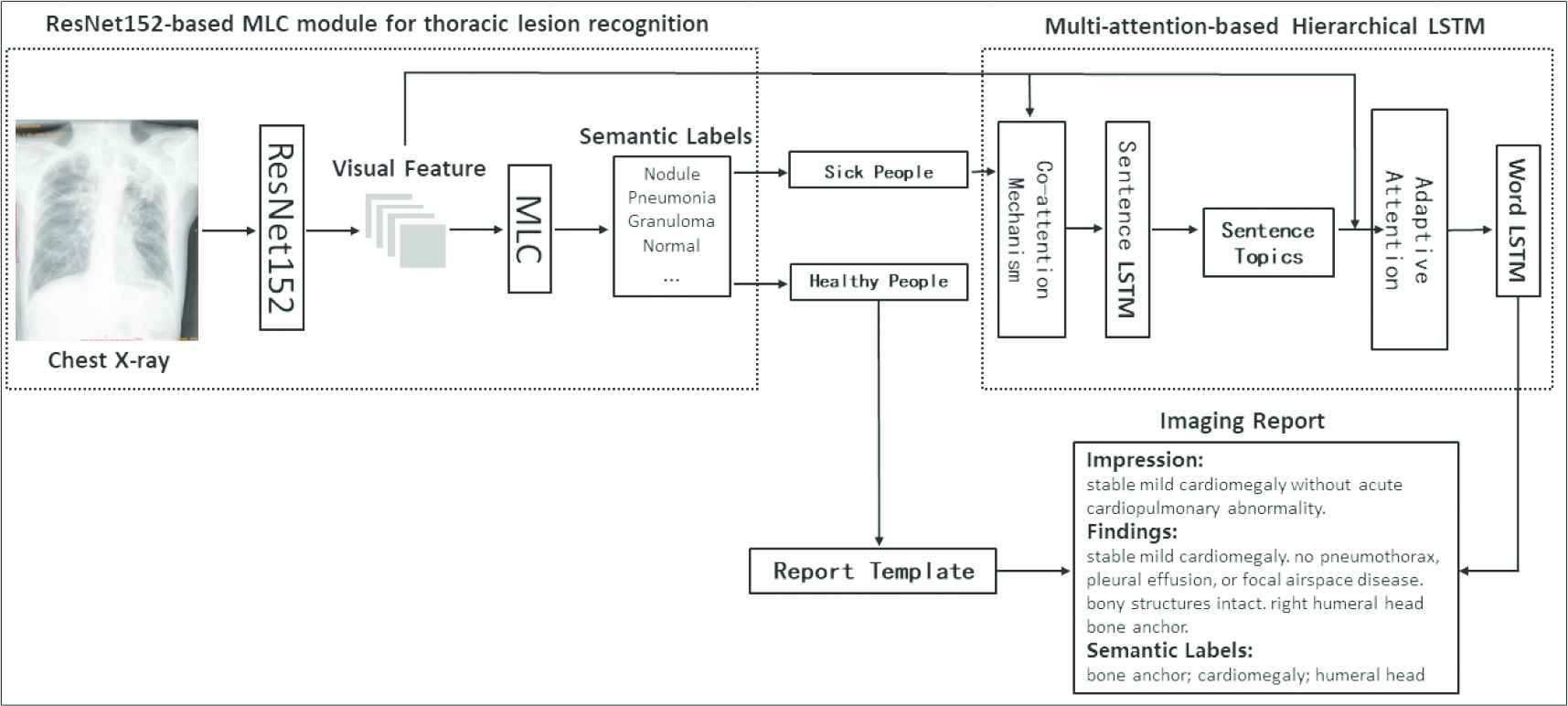
Over view of the framework of the proposed template-based multi-attention report generation model (TMRGM).
3.1. CNN-Based Thoracic Lesion Recognition
We define the identification of thoracic lesions as a MLC problem. Given a chest X-ray image, we first extracted the image feature V automatically using the ResNet152 model. Then we predict the probability distribution of 587 semantic labels collected from the IU X-ray dataset [18] via a MLC module
3.2. Chest X-Ray Image Classification
Considering the difference between descriptions from normal/abnormal reports, the TMRGM model first determines whether the given medical image belongs to healthy individuals or abnormal ones, and then utilizes different methods to generate reports for these two types of images. According to the distribution of semantic labels predicted by the thoracic lesion recognition model, we classify chest X-ray images based on the MLC module. We defined the image category as C, and the semantic label with the highest probability was Lmax, the image classification criteria was as follows:
In the formula (1), number 1 represents images from healthy individuals and number 0 represents images from abnormal individuals.
3.3. Template-Based Report Generation for Healthy Individuals
For healthy individuals, radiologists confirm no abnormalities and depict the normal organ or tissue with similar descriptions. In view of this, we constructed a library of chest X-ray report template for generating normal reports of healthy individuals. We first selected all the imaging reports of healthy ones from the IU X-ray dataset, and then we respectively collected sentences from two text field, “Findings” and “Impression.” Since many sentences in imaging reports express similar medical meaning (e.g., “pulmonary vascularity is within normal limits” and “pulmonary vascularity is normal”), we sorted these sentences according to their frequency in corresponding field and manually classified and labeled them. Specifically, first, we combined identical sentences (maybe some words have different singular or plural forms or tenses) into a single sentence. Second, we categorized sentences that have similar medical meaning. Third, we annotated the key words in each sentence for further analysis. Forth, we ranked the categories according to the sum of sentence frequency in each category, and selected one representative sentence from each category to construct a normal template library. Fifth, on average, the “Findings” field contains 3.4 sentences and the “Impression” field contains 1.5 sentences, we chose the top4 categories from “Findings” and the top2 categories from “Impression.” Finally, we use the representative sentences from the chosen 6 categories as the template sentences for generating normal imaging reports of healthy individuals.
3.4. Multi-Attention-Based Report Generation for Abnormal Individuals
3.4.1. Co-attention-based multimodal feature fusion
To better interpret abnormal findings, it is necessary to combine the local image features with high-level thoracic lesion labels. We employed the co-attention mechanism to fuse the image features extracted by ResNet152 and the text features of the thoracic lesion labels predicted by the MLC module. The feature fusion model assigned corresponding weights to different image regions while generating sentences, so that it can focus on the related image region and the thoracic lesion labels.
In particular, we first define a sentence LSTM model. At time t, let the image feature as
In formula (2),
3.4.2. Sentence topic generation based on sentence LSTM
The sentence LSTM contains three parts: (1) a single-layer LSTM network, which generates the LSTM hidden state
3.4.3. Adaptive attention-based word LSTM for sentence generation
There are many nonvisual words in the report context, such as “evidence,” “of,” “acute,” and “remain,” which cannot be aligned directly to a specific image region. Otherwise, in the training process, the gradient of nonvisual words will influence the alignment accuracy between visual words and image features. Therefore, we used the adaptive attention-based word LSTM model to generate sentences. During the process of word generation, the adaptive attention mechanism decides whether to use the image feature, the sentence topic, or rather the context feature to generate the current word. Figure 2 shows the structure of the word LSTM model based on the adaptive attention mechanism.

The structure of the word LSTM model based on the adaptive attention mechanism.
The adaptive attention mechanism [34] is an extension of the soft attention model proposed by Xu et al. [35]. As shown in the formula (13) and (14), at timestamp t, the adaptive attention mechanism assigns weights
The adaptive attention also improves the LSTM by introducing a new sentinel gate
To compute
4. EXPERIMENTS
4.1. Preprocessing of Chest X-Ray Dataset
Indiana University chest X-ray collection [18] is a public dataset containing 7470 chest X-ray images and 3955 de-identified radiology reports, and is commonly used for assessing imaging report generation models. Each report is comprised of several sections: Impression, Findings, and Indication, etc. We select Findings and Impression from the reports as our experimental data. The semantic labels annotated by MTI tools [36] are also collected for thoracic lesion recognition.
During the preprocessing stage, we resized all chest X-ray images into 224*224 pixels as the unified input of CNN model. The image quality is quite acceptable and we did not use additional data augmentation technologies. For collected MTI labels, we removed duplicates, lowercased all words, and obtained a set of 587 semantic labels. For texts extracted from Findings and Impression, we performed sentence segmentation, lowercased, delimitated punctuations, special characters, and extra spaces, and then converted numbers into a unified identifier “num.” Further, we constructed a dictionary base on the word frequency higher than 5 in imaging reports, in which 1173 words were included.
Figure 3 shows a processed chest X-ray report sample, including a chest X-ray image, corresponding semantic labels and textual descriptions.

A sample of processed chest X-ray report sample.
We filtered out 298 reports without MTI labels, and collected the rest of 3657 reports together with 6909 X-ray images as our experimental dataset. We divided the whole dataset into three parts, i.e., a validation set containing 500 randomly selected X-ray images, a test set containing another 500 images, and a training set containing the rest of 5909 images.
4.2. Experimental Settings
4.2.1. Implementation details
We carried out experiments on Windows Sever 2012 R2, Intel(R) Xeon(R) Gold 6130 64 CPU, 512GB memory, NVIDIA Tesla P100 16GB * 4 GPUs. The codes of TMRGM are implemented under the PyTorch framework and are available at https://github.com/546492928/TMRGM.
During the training process, the dimensions of hidden states in sentence LSTM and word LSTM are set to 512. The dimension of thoracic lesion word embedding, sentence topic embedding and report word embedding are also set as 512. We adopt a pretrained ResNet152 as image encoder, which is fine-tuned on the training set for obtaining chest X-ray image features. For the thoracic lesion MLC module, the visual features are 2048 dimensions extracted from the last average polling layer of ResNet152. For the multi-attention-based report generation module, visual features are extracted from the last convolutional layer, which yields a 7*7*2048 feature map. We use Adam optimizer with the initial learning rate of 0.0003 (dynamically reduced by 10% while the training error stop descending in 10 epochs), and the batch size is set as 16.
4.2.2. Evaluation metrics
We evaluated each submodule of our proposed method on different evaluation metrics. For evaluating the performance of MLC of thoracic lesions, we calculate precision (P), recall (R), F1 score, Recall@5, Recall@10, and Recall@20. Specifically, recall@N compares the number of correct labels in the top N predictions with the total number of labels in ground truth. For the chest X-ray image classification module, we calculate accuracy, specificity, and sensitivity. As to the imaging report generation module, we obtained BLEU [49], METEOR [50], ROUGE [51], and CIDEr [52] by the standard image captioning evaluation tool [53], which are commonly used in the field of machine translation and image captioning.
4.2.3. Comparison methods
For thoracic lesion recognition and chest X-ray classification, we compare the influence of different image encoders on the classification models. As a comparison experiment, we simultaneously built multiple CNN models such as VGG19 [37], Densenet121 [38], SENet154 [39], and ResNet152 [31] to extract visual features, as shown in Tables 1 and 3.
| Methods | P | R | F1 Score | Recall@5 | Recall@10 | Recall@20 |
|---|---|---|---|---|---|---|
| ResNet152 | 0.112 | 0.698 | 0.181 | 0.605 | 0.698 | 0.767 |
| VGG19 | 0.091 | 0.618 | 0.150 | 0.560 | 0.618 | 0.635 |
| Densenet121 | 0.112 | 0.682 | 0.180 | 0.595 | 0.682 | 0.756 |
| SENet154 | 0.112 | 0.701 | 0.180 | 0.602 | 0.701 | 0.775 |
| ResNet152 (top2) | 0.311 | 0.488 | 0.355 | __ | __ | __ |
Results of multi-label classification model based on different CNN models.
For chest X-ray report generation, we compare our proposed method with state-of-the-art method: TieNet [28], CoAtt [29], and Adapt-Att [34]. We also report TMRGM without introducing template. Further, we perform a qualitative assessment of the generated radiology reports manually.
4.3. Results
4.3.1. Results of thoracic lesion recognition
Table 1 shows the experimental results of thoracic lesion recognition based on different MLC models. It can be seen that the ResNet152-based MLC module achieved the precision of 0.112, the recall@5 of 0.605, the recall@10 of 0.698, and the F1 Score of 0.181, which outperform other methods. However, the best precision was only 0.112. For one thing, the 587 semantic labels increased the difficulty of building high-precision classifiers, while the training set only contains 5909 chest X-ray images. For another thing, the distribution of the semantic labels showed that each image in the training set contains two labels on average, which implies the confliction of our strategy on label selection (top 10). We tried to select the top2 labels predicted by ResNet152 as a comparison, and achieved a precision of 0.311, a recall of 0.488, and a F1 score of 0.355.
Table 2 shows two examples of ResNet152-based MLC module for thoracic lesion recognition. For the first image, the model correctly identified three semantic labels, namely atelectases, atelectasis, and opacity. According to these three lesions, patients can go to the respiratory department for medical treatment. As to the other one, we recognized a lesion “cardiomegaly,” which reminds the patient to see the cardiologist.
| Chest X-ray Image | Predicted Labels | MTI Labels |
|---|---|---|
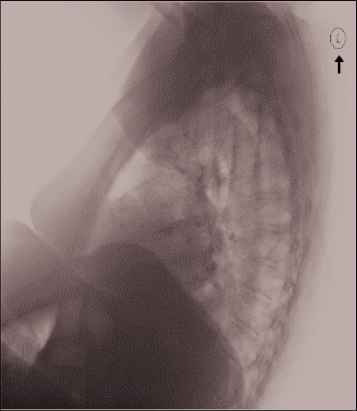 |
atelectases; atelectasis; opacity; cardiomegaly; scarring; degenerative change; calcified granuloma; normal; pleural effusion; granuloma |
atelectases; opacity; atelectasis; hiatal hernia; infection |
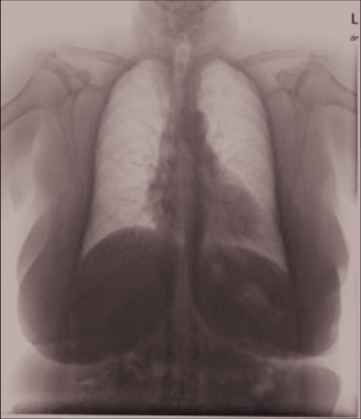 |
cardiomegaly; degenerative change; opacity; atelectases; atelectasis; scarring; normal; calcified granuloma; granuloma; pleural effusion |
cardiomegaly |
Examples of ResNet152-based MLC module for thoracic lesion recognition.
4.3.2. Results of chest X-ray image classification
According to whether the “Normal” label achieves the highest probability in predicted labels, we classified chest X-ray images into healthy individuals and abnormal ones. We compare the ResNet152-based classification module with other CNN-based binary classification models, such as VGG19, Densenet121, SENet154, and Inception-V3 [40]. Table 3 shows the experimental results of chest X-ray image classification. The ResNet152-based classification model achieved the best accuracy of 0.73, the DenseNet121 achieved the best specificity of 0.803, and the SENet achieved the best sensitivity of 0.758. The ResNet152 achieved the best 95% confidence interval of the accuracy ([0.691, 0.769]), followed by the Densenet121 ([0.674, 0.754]) and the SENet154 ([0.672, 0.752]). In the ResNet152-based classification model, error case study reveals that a part of normal cases were misclassified as abnormal ones. A statistical analysis reveals that the ratio of images from healthy and abnormal individuals in the training set was about 2:3, which indicates that the performance of chest X-ray image classification is to some extent affected by the data imbalance.
| Method | Accuracy | Confidence Interval (95%) | TP | TN | FP | FN | Specificity | Sensitivity |
|---|---|---|---|---|---|---|---|---|
| ResNet152 | 0.73 | [0.691, 0.769] | 137 | 228 | 61 | 74 | 0.789 | 0.649 |
| VGG19 | 0.578 | [0.535, 0.621] | 289 | 0 | 211 | 0 | 0 | 1 |
| Densenet121 | 0.714 | [0.674, 0.754] | 125 | 232 | 57 | 86 | 0.803 | 0.592 |
| SENet154 | 0.712 | [0.672, 0.752] | 160 | 196 | 93 | 51 | 0.678 | 0.758 |
| Inception-V3 | 0.708 | [0.668, 0.748] | 128 | 226 | 63 | 83 | 0.782 | 0.607 |
Results of chest X-ray classification.
4.3.3. Templates for chest X-ray report generation
For generating the reports of healthy individuals, we manually constructed templates based on the Findings and Impression text respectively. Specifically, the Impression section contains 63 subclasses of the report sentence (partly shown in Table 4), and the “Findings” field contains 150 subclasses (partly shown in Table 5). According to the sum of the sentence frequency in each subclass, we selected the top2 high frequency subclass for the Impression and the top4 subclass for the Findings. Then the combination of the representative sentences from the selected six subclasses forms a complete Chest X-ray report template (see Table 6).
| Class | Representative Sentence | Frequency | Key Words |
|---|---|---|---|
| 1 | No acute cardiopulmonary abnormality | 817 | Cardiopulmonary abnormality |
| 2 | No active disease | 384 | Abnormality |
| 3 | Heart size is normal and lungs are clear | 76 | Heart size; lung |
| 4 | The heart size and cardio mediastinal silhouette are within normal limits | 67 | Heart size; cardio mediastinal silhouette |
| 5 | No acute pulmonary disease | 55 | Pulmonary disease |
Some part of manually annotated sentences in the Impression section.
| Class | Representative Sentence | Frequency | Key Words |
|---|---|---|---|
| 1 | No focal consolidation pleural effusion or pneumothorax | 579 | Pleural effusion; pneumothorax |
| 2 | The lungs are clear | 550 | Lung |
| 3 | The cardiomediastinal silhouette is within normal limits | 320 | Cardiomediastinal silhouette |
| 4 | The heart is normal in size | 315 | Heart size |
| 5 | Visualized osseous structures of the thorax are without acute abnormality | 163 | Thorax; osseous structure |
Some part of manually anotated sentences in the Findings section.
| Section | Template |
|---|---|
| Impression | No acute cardiopulmonary abnormality No active disease |
| Findings | No focal consolidation pleural effusion or pneumothorax The lungs are clear The cardiomediastinal silhouette is within normal limits The heart is normal in size. |
The complete template of chest X-ray reports of healthy individuals.
4.3.4. Results of chest X-ray report generation
Table 7 shows results of Chest X-ray report generation on the automatic metrics. The evaluation metrics, such as BLEU score, METEOR, ROUGE, and CIDEr, are based on n-gram similarity between the generated sentences and the ground-truth sentences. The difference between these metrics lies in the various strategies of n-gram similarity calculation and weight assignment. We compared our proposed TMRGM model with three state-of-the-art methods based on the test set, as shown in Table 7, which demonstrate the comparable performance of TMRGEM to the SOTA. The Adapt-att represents the hierarchical LSTM model based solely on multi-attention mechanism, which achieved the best ROUGE of 0.316 and the CIDEr of 0.387, suggesting that the hierarchical model is better for modeling paragraphs. Our TMRGM model obtained the preferable BLEU scores and the METEOR of 0.183, which indicates the high semantic similarity between generated report sentences and the ground-truth sentences. By comparing the results of TMRGM model and TMRGM without templates, we can see that the introduction of chest X-ray report template can improve the BLEU scores and METEOR, suggesting that that the template-based report generation is linguistically in line with the reports of healthy individuals.
| Method | BLEU-1 | BLEU-2 | BLEU-3 | BLEU-4 | METEOR | ROUGE | CIDEr |
|---|---|---|---|---|---|---|---|
| TieNet [28] | 0.286 | 0.160 | 0.104 | 0.074 | 0.108 | 0.226 | -- |
| CoAtt (Jing et al., 2018) | 0.303 | 0.181 | 0.121 | 0.084 | 0.132 | 0.249 | 0.175 |
| Adapt-att | 0.378 | 0.255 | 0.185 | 0.138 | 0.162 | 0.316 | 0.387 |
| TMRGM(without template) | 0.380 | 0.259 | 0.188 | 0.141 | 0.163 | 0.317 | 0.391 |
| TMRGM | 0.419 | 0.281 | 0.201 | 0.145 | 0.183 | 0.280 | 0.359 |
Result of chest X-ray report generation.
4.3.5. Qualitative analysis
In this section, we perform the qualitative analysis on the generated reports. Table 8 presents two abnormal cases of chest X-ray reports generated by the TMRGM model and Table 9 shows an example of template-based reports generated for healthy ones.
| Chest X-ray | Generated Report | Ground Truth |
|---|---|---|
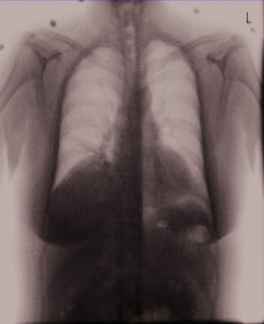 |
No acute cardiopulmonary abnormality. The heart size and pulmonary vascularity appear within normal limits. The lungs are free of focal airspace disease. No pleural effusion or pneumothorax is seen. |
Right middle lobe airspace disease may reflect atelectasis or pneumonia. The cardiomediastinal silhouette is normal size and configuration. Pulmonary vasculature within normal limits. There is right middle lobe airspace disease may reflect atelectasis or pneumonia. No pleural effusion. no pneumothorax |
 |
Stable cardiomegaly with prominent perihilar opacities which may represent scarring or edema. There is stable cardiomegaly. there is no pneumothorax. |
Findings concerning for interstitial edema or infection. Heart size is mildly enlarged. There are diffusely increased interstitial opacities bilaterally. No focal consolidation pneumothorax or pleural effusion. No acute bony abnormality. |
Examples of generated chest X-ray reports for abnormal individuals.
| Chest X-ray | Generated Report | Ground Truth |
|---|---|---|
 |
No acute cardiopulmonary abnormality. No active disease. No pneumothorax or pleural effusion. The lungs are clear. The cardiomediastinal silhouette is within normal limits. The heart is normal in size. |
No acute cardiopulmonary findings. No focal consolidation. No visualized pneumothorax. No pleural effusions. Heart size normal. The cardiomediastinal silhouette is unremarkable |
An example of generated chest X-ray reports for healthy individuals.
As shown in Table 8, for the upper case, two sentences of normal descriptions are semantically similar with the ground-truth sentence, such as “pulmonary vascularity appear within normal limits.” versus “pulmonary vasculature within normal limits”; and “no pleural effusion or pneumothorax is seen.” versus “no pleural effusion. no pneumothorax.” As to the second case in Table 8, the TMRGM model performs acceptable on generating abnormal descriptions of chest X-rays, e.g., the predicted sentence “stable cardiomegaly with prominent perihilar opacities which may represent scarring or edema,” is semantically similar with the real sentence “findings concerning for interstitial edema or infection. heart size is mildly enlarged. there are diffusely increased interstitial opacities bilaterally.”
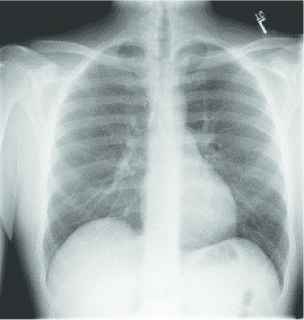
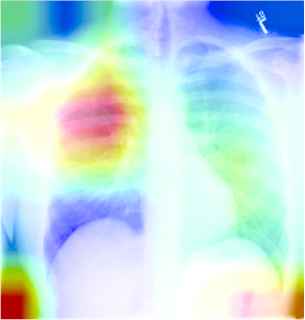
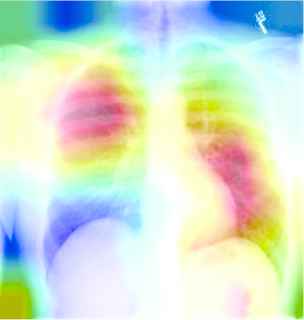
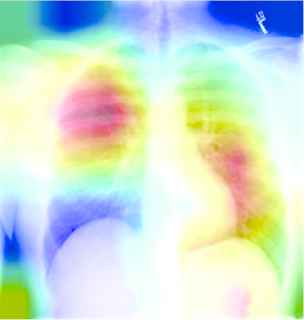
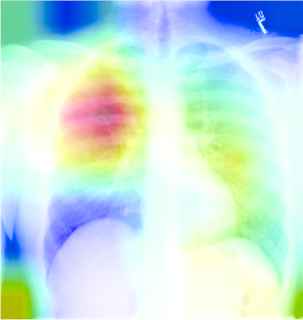
Table 9 described the chest X-ray of a healthy individual from several aspects, such as the cardiopulmonary function (“no acute cardiopulmonary abnormality”), the pleural lesions (“no pneumothorax or pleural effusion”), the costal mediastinum outline (“the cardiomediastinal silhouette is within normal limits”), the cardiac shape and size (“the heart is normal in size”). It can be observed that the descriptions of multiple anatomic structures are grammatically and logically in accord with the ground-truth sentences, which demonstrate the chest X-ray report template is highly similar with the real normal reports in the OpenI IU X-ray dataset. As shown in Table 10, the visualization heat map reveals the attentive image region while generating a specific sentence. The highlights in the heat map represent the image features used to generate the corresponding sentence, and the darker the color, the greater the weight. However, it is hard to explain the correlation between generated sentences and image features.
 |
 |
 |
 |
| Generated report | No acute cardiopulmonary findings | No acute cardiopulmonary findings | The lungs and pleural spaces show no acuteabnormality |
 |
 |
 |
|
| The cardiomediastinal silhouette and pulmonary vasculature are within normal limits in size | no typical findings of pulmonary edema | no typical findings of pulmonary edema | |
| Ground truth | Negative for acute abnormality. The cardiomediastinal silhouette is normal in size and contour. no focal consolidation pneumothorax or large pleural effusion. normal xxxx. |
||
A visualization example of generated sentences and corresponding heat map.
5. DISCUSSION
Automatic chest X-ray report generation will facilitate radiologists to improve the efficiency of diagnosis and report writing. The proposed TMRGM model achieved comparable performance with SOTA models on chest X-ray report generation. However, it is still far from clinical usage in realistic scenarios.
First, in the training phase, we collected semantic labels and built report templates entirely based on the training reports from the IU X-ray. Then we test our proposed model based on another 500 samples. We found that in generated reports of abnormal individuals, most sentences are normal descriptions, while the proportion of abnormal descriptions is relatively small. This problem may due to the imbalance of normal and abnormal descriptions in the training set (in the IU X-ray dataset, each report contains 3.7 normal sentences and 2.6 abnormal sentences on average). Empirically, the data scale, completeness, normalization, and quality of imaging reports are important factors for training. One further improvement is introducing high-quality parallel datasets, such as the recently released MIMIC-CXR dataset, so as to train the model better. It is also necessary for us to validate the generalization performance on external data source.
Second, unlike common natural images, the difference of visual features in medical images is not obvious, and the ambiguous situations are quite often, such as the same disease with diverse visual features, or the similar image features attributed to different diseases. The TMRGM model extracted image features based on the ResNet152, and involved the co-attention as well as the adaptive attention mechanism. The introduction of the adaptive attention mechanism chooses reasonable features for generating different kinds of words, which to some extent, alleviates the problem of unaligned non-visual words and image features. However, by reviewing the visualization heat maps of TMRGM, we found it is hard to explain the correlation between generated sentences and image features. One optimizing strategy is to segment chest X-ray images by referring to the description sequence and body parts specified in reports, and then extract local image features respectively. Since each body parts has specific semantic labels, the problem of image feature extraction and classification would be more simplified. Another direction for improvement is to explore emerging explainable deep learning networks, combining with state-of-the-art data augmentation for better understanding and interpreting radiology images.
Third, we selected the top 10 semantic labels from the MLC module as the thoracic lesions. Based on this rule, we achieved high recall but poor precision on thoracic lesion recognition. It is necessary to explore more reasonable label selection strategies. In addition, in view of the increasing open access Covid-19 dataset, our method can be further optimized for assisting the current Covid-19 diagnosis, such as identifying thoracic lesions and automatically writing radiology reports, and reduce the workload of doctors.
Fourth, the dictionary used by the TMRGM model to generate the medical imaging report contains anatomical locations like right, left, upper, and lower. However, due to the uneven distribution of words in the training set and the low frequency of anatomical locations, most of the generated reports do not contain accurate anatomical locations. This is also a limitation of this study. In further research, we will focus more on the location of the disease in the medical imaging pictures and how to accurately generate the description of the anatomical locations.
6. CONCLUSION
In this paper, based on a systematic review of thoracic lesion recognition and medical imaging report generation, we proposed a template-based multi-attention model (TMRGM) for automatically generating reports of chest X-rays. By exploring the linguistic characteristics of report texts, we implemented different report generation methods for healthy individuals and abnormal ones respectively, and validate the effectiveness of TMRGM based on the IU X-ray dataset. It is helpful for radiologists to quickly identify the thoracic lesions and write high-quality chest X-ray reports. That facilitates the daily work of medical imaging examination and reduce their burden of image reading and report writing.
CONFLICT OF INTEREST
The authors declare they have no conflicts of interest.
AUTHORS' CONTRIBUTIONS
All authors have made significant contributions to the manuscript including its conception and design, the analysis of the data, and the writing of the manuscript. All authors have reviewed all parts of the manuscript, take responsibility for its content, and approve its publication.
ACKNOWLEDGMENTS
This work has been supported by the National Natural Science Foundation of China (Grant No. 61906214), the Beijing Natural Science Foundation (Grant No. Z200016), CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant No. 2018-I2M-AI-016), and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (Grant No. 2018PT33024).
REFERENCES
Cite this article
TY - JOUR AU - Xuwen Wang AU - Yu Zhang AU - Zhen Guo AU - Jiao Li PY - 2021 DA - 2021/05/05 TI - TMRGM: A Template-Based Multi-Attention Model for X-Ray Imaging Report Generation JO - Journal of Artificial Intelligence for Medical Sciences SP - 21 EP - 32 VL - 2 IS - 1-2 SN - 2666-1470 UR - https://doi.org/10.2991/jaims.d.210428.002 DO - 10.2991/jaims.d.210428.002 ID - Wang2021 ER -
