Toward Artificial Intelligence by using DNA molecules
- DOI
- 10.2991/jrnal.2018.5.2.12How to use a DOI?
- Keywords
- DNA computing; Molecular Robotics; Strand Displacement Reactions
- Abstract
Molecule Robotics have been realized by using bio molecules such as DNA, proteins, for example in Molecular Robotics Research Project [2012–17, JSPS]; bio molecules, such as DNA or proteins are highly structured and they already have a kind of “intelligence” and adapt to environment change. Hence, we have tried to extract their “ability” and induce Intelligence artificially. We have used well-known DNA reactions, Seesaw gate reaction and we found that DNA molecule can sense concentration of single strand input sequence or quasi-input (including mismatched base pairs and point mutations in the sequence) and chooses higher concentration one; this result shows DNA molecule (short DNA sequence) can adapt to its environment.
- Copyright
- Copyright © 2018, the Authors. Published by Atlantis Press.
- Open Access
- This is an open access article under the CC BY-NC license (http://creativecommons.org/licences/by-nc/4.0/).
1. Introduction
Adelman solved Hamilton Path Problem by using a computer, which was synthesized by DNA molecules. His research opens up novel era of molecular computing. DNA computing has evolved as a method of Bonino technology and realized a molecular robot.
For example, S. Nomura’s team has chemically implemented a molecular robot [1]. This robot is composed of bio molecules, which are composed of a body, an actuator, and an actuator-controlling device (clutch); whose body is composed of a lipid bilayer, the actuator consists of proteins including kinesin, and microtubules and the clutch made of designed DNA molecules. This clutch transmits the generated force from motor proteins to the membrane and the shape of the body changes. Gears of the clutch are changed by a signal molecule composed of sequence-designed DNA.
2. Molecular Artificial Intelligence
Artificial Intelligence, AI has developed on electrical device, where all information is coded as electrical signals. On the other hand, in a molecular robot all information is coded as chemicals such as DNA or proteins. Hence, if we can realize Artificial Intelligence by using chemicals, we will be able to give intelligence to the molecular robots.
2.1. Abstract Chemical Reactions by DNA
Several logical gates have been realized by using DNA [4]. Various simulations of Abstract Chemical Reactions, ACR based on Chemical kinetic have been made by using the DNA logical gates [3]. For example, Fujii et. al. [5] chemically implemented the Lotoka-Vorterra, LV model and showed that their reaction can realize the same behavior to analytic solution of LV model by differential equations.
Quian et.al., have proposed collection of DNA reactions as a “toolbox” to implement ACR by using DNA; they showed that an Artificial Neural Network can be implemented by using the toolbox [3].
3. Intelligence of Molecular Robot
Philosopher, prof. Kazuhisa Todayama has argued a possible development of philosophy as Concept Engineering. Concept engineering provides novel concept based on thinkers’ assets. For example, in the case when we have to face difficult problems such as creating a regulation for robots in order to ready for a case when a fully autonomous intelligent robot kills a human. In such a case, philosophers can provide wide varieties of concepts from different aspects and they can support to create a novel concept through collaborating with specialists and citizens. Todayama points out that Human right is a legacy of concept engineering, which has been invented by philosophers based on the asset of philosophy [2,3].
3.1. How to create intelligence in molecular robots
Todayama proposed how to create intelligence in a molecular robot from concept engineering view point; if we let the molecular robot free as possible as we can and if the robot can have own purpose, intelligence will be emerged, where any purpose is granted and no matter how it is simple or not, it is important to have its own purpose. In order to create molecular AI, we give a purpose to a robot, which is maintaining its self.
4. Methods
Chemical reactions in the toolbox are mainly modification of the toehold strand displacement reaction of DNA. Strand displacement reaction is initiated from complementary single-stranded domains and branch migration process exchanges a single strand DNA with double strand DNA through random-walk like; the complementary single-stranded domains are called as toeholds. In this reaction, at least one single stranded nucleic acid is released and this released strand can evoke downstream reactions. Since branch migration does not change the free energy during the reaction of exchanging strand pairs, it can be described by a random walk process with equal transitions probabilities of displacement in both directions at each position of the branch point [5].
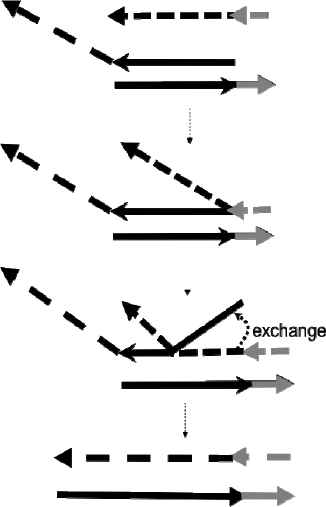
Strand displacement reaction; In this example, the short strand, which is denoted as an arrow binds to the toehold domains, through branch migration process across region 2, the strand is displaced and freed from the complex.
In order to implement molecular AI, we modified the Seesaw gate reaction [4]; since this reaction is self-maintaining reaction, we can give the reactions a purpose as self-maintaining. These reactions are composed of basically two strand displacement reactions; input + gate:fuel ↔ gate:input + fuel (1) and gate:output + input ↔ gate:input + output (2); in order to confirm the reactions circulate, we used the following reaction; fuel + gate(Quencher):output input(Fluorescence) ↔ gate(Quencher):input(Fluorescence) + output + gate(Quencher):fuel (3).
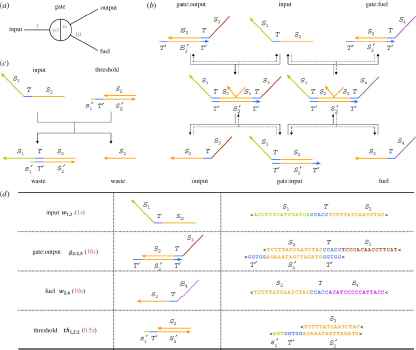
Seesaw gate reaction [4]: (a) Abstract gate diagram. Red numbers indicate initial concentrations. (b) The DNA gate motif and reaction mechanism. S1, S2, S3 and S4 are the recognition domains; T is the toehold domain; T0 is the Watson–Crick complement of T, etc. Arrowheads mark the 30 ends of strands. Signal strands are named by their domains from 30 to 50, All reactions are reversible and unbiased; solid lines indicate the dominant flows for the initial concentrations shown in (a), while the reverse reactions are dotted. (c)
5. Result
From biochemical experiments, we observed that when we added non-mutated input and mutated input, the system selected higher concentration one (Fig.3). And we also confirmed the behaviors in the time series of the concentration of normal input and mutated input sequences showed oscillations (Fig.3). And when the concentration decreased, the concentration of another input increased, hence we conjecture that the system selects higher concentration one because the selected input sequences were switched among non-mutated and mutated input sequences. These results illustrate that the system made of DNA reaction networks is able to select input sequences in higher concentration so it will be able to adapt to the environmental change. Therefore, from biological and philosophical points of views, this system realizes molecular Artificial Intelligence (Fig.3).
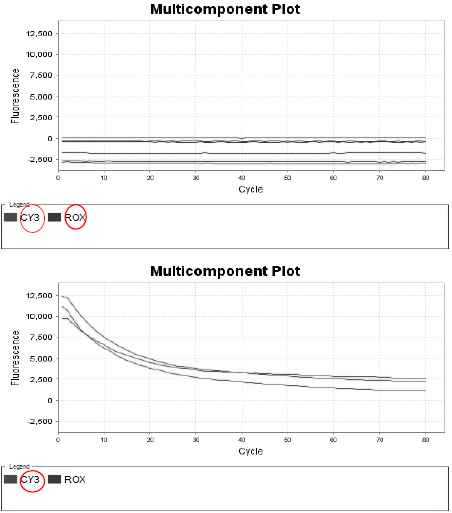
Time series of concentration of Input sequences obtained by the real time PCR: (Upper) No input sequence, (Lower) Input sequences without toehold, the concentration of input sequences decreased through binding to gate:output sequences. CY3 and ROX are fluorescent dyes to label DNA sequences.
Acknowledgement
Prof. Ken Komiya (Tokyo Institute of Tech.) gives insightful comments and suggestions. This research was supported by KAKENHI Grant No. 24104003 (JSPS) and 16H03093 (MEXT).
References
Cite this article
TY - JOUR AU - Yasuhiro Suzuki AU - Rie Taniguchi PY - 2018 DA - 2018/09/30 TI - Toward Artificial Intelligence by using DNA molecules JO - Journal of Robotics, Networking and Artificial Life SP - 128 EP - 130 VL - 5 IS - 2 SN - 2352-6386 UR - https://doi.org/10.2991/jrnal.2018.5.2.12 DO - 10.2991/jrnal.2018.5.2.12 ID - Suzuki2018 ER -
