Characterizations of a Food Decapeptide Chelating with Zn(II)
- DOI
- 10.2991/efood.k.200727.001How to use a DOI?
- Keywords
- Peptide; zinc; binding sites; molecular docking; structure
- Abstract
Walnut proteins and peptides have been reported to have great zinc-carrying activity. In this study, the decapeptide EPNGLLLPQY (WP-10) derived from walnut protein has been prepared to figure out zinc binding mechanisms and structure–activity relationships. The space-conformation of this peptide has converted after being chelated with Zn(II). As to secondary structure, the β-sheet structure of the peptide turned into α-helix and β-turn structure. According to the atomic absorption spectra analysis, one peptide could bind with one Zn(II) via four different binding sites. The signals of different groups in infrared spectroscopy have shifted before and after chelation, and the major functional groups of this peptide involved in chelation are –NH2 and –C=O. GLU1, PRO2, ASP3, GLU4, and LEU6, and the molecular docking proved their participation in the chelating with Zn(II).
- Copyright
- © 2020 International Association of Dietetic Nutrition and Safety. Publishing services by Atlantis Press International B.V.
- Open Access
- This is an open access article distributed under the CC BY-NC 4.0 license (http://creativecommons.org/licenses/by-nc/4.0/).
1. INTRODUCTION
Zinc is an important mineral that is involved in physiological metabolism, and functions in more than 300 metalloenzymes as a catalytic component ensuring the structural integrity of proteins [1]. Moreover, zinc has a vital influence on the immune system, as it is required for the activation of different kinds of immune cells [2]. Besides, zinc plays an important role in maintaining cell reproduction, gene expression, regulatory function, neurogenesis and neurotransmission [3,4]. But zinc cannot be synthesized in the body, and human zinc reserves are limited. Therefore, biological zinc must be continuously supplied from the individuals diet [5]. Zinc deficiency can seriously affect the immune system and destroy the redox balance of the body, leading to immune dysfunction, neurological decline, and the occurrence of some pathological diseases [6].
At present, zinc can be obtained from mineral salts and metal chelating agents [7]. However, mineral salts have poor bioavailability due to the inhibition of dietary components such as tannins, phytates, and dietary fibers [8,9]. Therefore, it is necessary to develop alternatives that can improve the bioavailability of zinc. Subsequently, more attention has been paid to the study of new supplements of zinc, such as zinc-binding proteins or peptides as functional food.
Pioneering studies have shown that expressed proteins bear metal-coordinating side chains or incorporate such groups [10], and the same applies to food-derived peptides. A great number of food-derived peptides with metal chelating abilities have been identified such as the sesame protein [11], whey protein [12], milk protein [13], Antarctic krill protein [14], amyloid protein [15] and walnut protein [16]. Furthermore, food-derived peptides can be easily digested and absorbed due to having a small molecular weight and a simple structure. However, the activity of the walnut-derived polypeptide with zinc chelation has been hardly reported.
The study presented here demonstrates the structure–activity relationship of Zn(II) and walnut-derived polypeptides share after chelation. In our team’s previous research, several peptides were isolated and identified from walnut protein. Among them, Glu-Pro-Asn-Gly-Leu-Leu-Leu-Pro-Gln-Tyr (WP-10) showed metal ion chelating activity. The aim of this study was to prepare zinc-chelating peptides (purified synthetic walnut-derived polypeptides) and provide the theoretical basis behind using zinc-chelating peptides from walnut protein as a functional food ingredient.
2. MATERIALS AND METHODS
2.1. Materials and Chemicals
The walnut peptide WP-10 was synthesized from Cellman Biotech Limited Corporation (Hefei, China). Zinc chloride buffer solutions of varying pH levels and all other reagents were purchased from Damao Chemical Reagent Co. Ltd. (Tianjin, China).
2.2. Preparation of the Zinc-Chelating Peptide
WP-10 (0.2 mmol/mL) was dissolved in a Tris-HCl buffer (0.05 mol/L, pH 5.0) containing ZnCl2. The centrifuge tube containing the mixture was placed in a constant temperature shaking water bath at 47°C for 80 min. After that, the free zinc ion was removed by dialysis for 24 h using a 100 Da molecular weight dialysis bag made of semi-permeable membrane, and collected retentates (containing chelated zinc). Additionally, a part of it was lyophilized as a complete chelate for the following study.
2.3. Zinc Chelating Capacity Analysis
The free zinc content in the supernatant was evaluated by flame atomic absorption spectrometry (ZA3000, HITACHI, Japan) according to Wu, Wenfei and Smichowski’s methods with some modifications [17,18], the chelated zinc content was calculated. The experiments were performed in triplicates and data was presented as the mean of each repeated experiment.
2.4. Ultraviolet–Visible Spectroscopy
In order to testify the occurrence of the zinc-chelating peptide reaction and investigate the functional mechanism between the peptide and peptide-zinc chelates, the absorbance values were recorded in the wavelength range from 200 to 1100 nm with an Ultraviolet–Visible (UV–Vis) spectrophotometer (F-2700, HITACHI, Japan). The zinc-chelating peptide and pure peptide were dissolved in a 0.1 mol/L Tris-HCl buffer (pH 7.4). Besides, samples were put in a quartz cuvette with a path length of 0.5 cm [19–21].
2.5. Circular Dichroism Spectroscopy
The secondary structure of the zinc-chelating peptide and pure peptide was investigated with a circular dichroism (CD) spectrophotometer (J-1500, Jasco, Japan) and the zinc-chelating peptide was compared to the pure peptide. The spectra of the zinc-chelating peptide and pure peptide were recorded in the wavelength range from 190 to 260 nm with a scanning rate of 20 nm/min, and a 1 nm bandwidth. In these experiments, the data was represented as the average of three independent scans with the control CD signal subtracted [22].
2.6. Fourier Transform Infrared Spectroscopy
The zinc-chelating peptide and pure peptide were inspected by Fourier Transform Infrared Spectroscopy (FTIR) (Frontier, Perkin Elmer, USA). One milligram of the sample was mixed with 100 mg of dried KBr, pulverized in an agate mortar, and then pressed into a transparent round shape [23]. The spectra were recorded in the wavenumber region of 4000–400 per cm with a resolution of 4 per cm and an average scan of 16 times [24]. The potential zinc-chelating sites were deduced from the significant shifts in absorption bands [16].
2.7. Molecular Dynamics Simulation
The discovery Studio 2017 R2 software with minor modifications was used to conduct molecular docking between WP-10 and zinc ions. The amino acid sequence of peptides was entered and the peptide structure was pretreated with the Prepare Protein tool. The structures of the ligands were generated by the DS 2017 R2 software and the Full Minimization tool was utilized to give the minimized energy. The peptide structure was processed with the clean and prepare protein program to model missing loop regions, remove water, add hydrogen, and optimize the bond length. The docking sites have a spherical center with a center diameter of 8 Å as the active site to cover the docking region. The docking program was carried out using the partial flexibility program CDOCKER protocol. Evaluation of molecular docking results was based on CDOCKER energy scores, interaction sites, and interaction force types [25,26].
3. RESULTS AND DISCUSSION
3.1. Zinc Chelating Capacity of Peptide
Zinc chelating capacity of peptide was shown in Figure 1. The amount of chelated Zn(II) on the WP-10 increased when the concentration of Zn(II) increased. The zinc chelating capacity did not increase significantly when zinc concentration reached 1.5 μmol/L. The reason may be that it is already saturated. Therefore, zinc ion concentration was one of the most important factors affecting the chelating capacity.

Effects of zinc ion concentration on zinc chelating capacity. The zinc chelating capacity was determined by flame atomic absorption spectrometry. All the samples were determined three times and the data were reported as mean ± SD.
3.2. Ultraviolet–Visible Spectroscopy Analysis
As the curve shown in Figure 2, there are peaks in 280 and 220 nm, respectively as determined by the intensity and dislocation change of UV–Vis spectroscopy. As we all know, Tyrosine (Tyr) can absorb ultraviolet light at 278 nm, Tryptophan (Trp) can absorb ultraviolet light at 279 nm, Phenylalanine (Phe) can absorb ultraviolet light at 259 nm. The peak detected in 280 nm was small, which was attributed to lack of Phe, Tyr and Trp. After chelating interactions, the peak value slightly reduced, which was explained by the facts that some of Tyr in WP-10 were buried after chelation with Zn(II). These results were corresponded to the results of Figure 1. As for the peaks in 220 nm, it also decreased after chelating. It indicated that WP-10 were chelated to Zn(II) and the content of free peptides was decreased.

UV–Vis spectrum of the decapeptide and zinc-chelating peptide. The WP-10 and zinc-chelating peptide were dissolved in Tris-HCl buffer (0.1 mol/L, pH 7.4). The spectra were recorded between 200 and 1100 nm with a scan speed of 20 nm/min.
3.3. Circular Dichroism Spectroscopy Analysis
In circular dichroism spectrum analysis, if a negative peak ranged between 190 and 220 nm, it indicates that the peptide has a random structure. Similar results were found as for β-sheet structure, when negative peak ranges from 210 to 225 nm. As shown in Figure 3, the curve of circular dichroism spectrum for WP-10 had a negative peak at 190–225 nm. Therefore, it may have β-sheet and random structures. Moreover, it could be found that WP-10 contains mostly the structure of random and β-sheet which was 79.6% and 20.4% in the peptide, respectively.
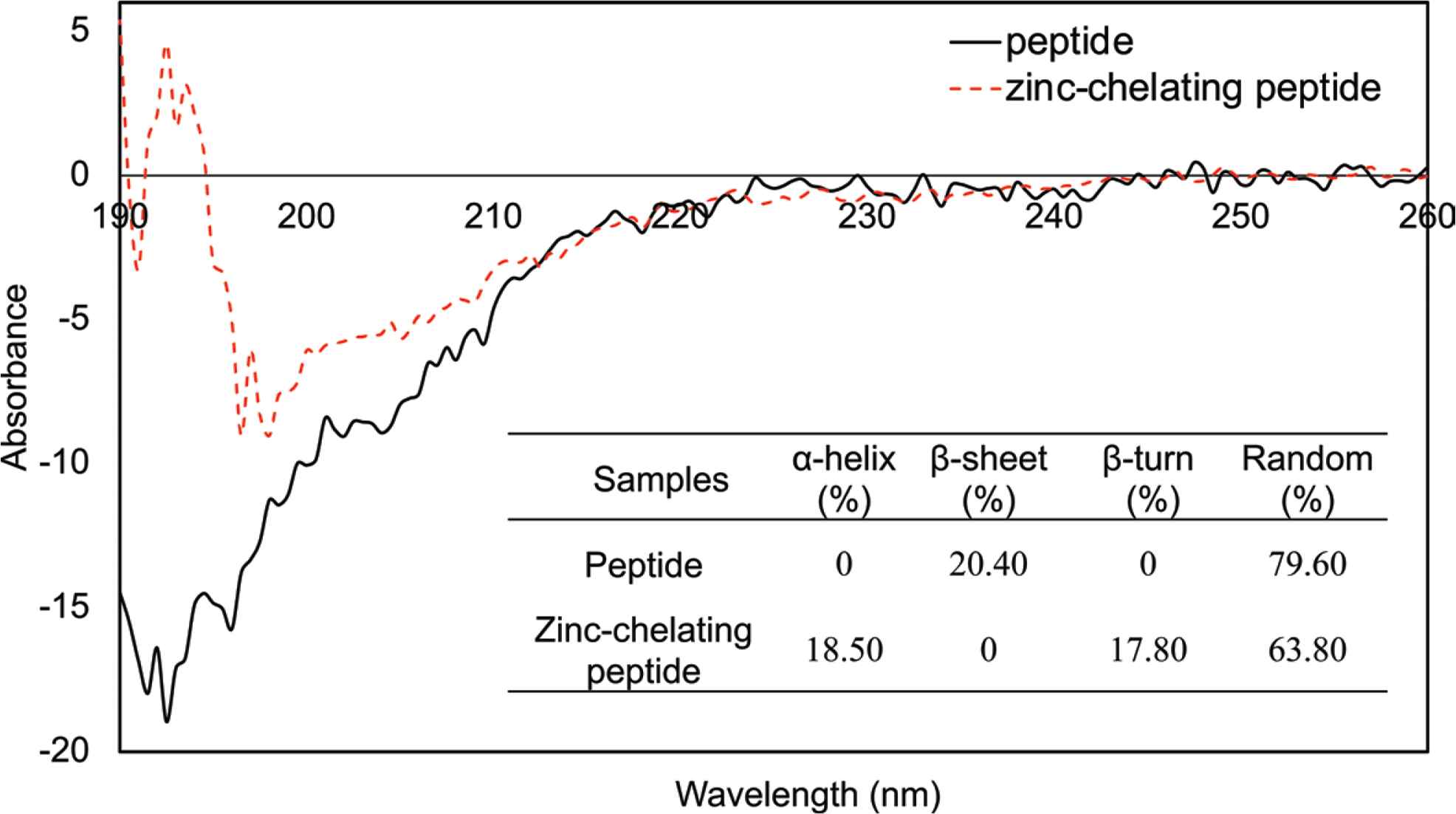
CD spectrum of the decapeptide and zinc-chelating peptide. The WP-10 and zinc-chelating peptide were dissolved in Milli-Q water to a final concentration of 0.20 mmol/mL. The spectra were recorded between 190 and 260 nm with a scan speed of 20 nm/min.
When the WP-10 was chelated with Zn(II), its molecular structure changed accordingly. It could be found that a positive peak ranged from 190 to 195 nm while a negative peak from 195 to 220 nm, which indicated that α-helix and β-turn presented after WP-10 chelated with Zn(II), while β-sheet disappeared. The proportions of α-helix, β-turn and random structures were 18.5%, 17.8%, and 63.8%, respectively.
Compared to these two results, it was verified that the effect of Zn(II) on the structure of WP-10 was consistent with the results of UV–Vis spectroscopy analysis. In a Zn(II) rich system, Zn(II) chelating WP-10 was a process that destroyed the β-sheet structure and reconstituted it to α-helix and β-turn in WP-10 resolution. In this process, a part of random structures was involved in this conversion.
3.4. Fourier Transform Infrared Spectroscopy Analysis
As shown in Figure 4, the wave number (WN) region of infrared spectrogram (IR) fingerprint spectrum was ranged from 650 to 1350 per cm. It could be found that the waveforms of WP-10 and zinc-chelating peptide complexes were significantly different while waveforms among the chelated peptides in different Zn(II) concentration were similar. These fingerprint spectrums could be used to distinguish this peptide whether chelated with Zn(II) or not. Combined with Figures 1 and 2, the IR and structure of the pure peptide, the peaks of functional groups, such as C–C(O), NH, RNH2, –CH, –CH3, C=O, could be identified. The amide-I vibration (C=O stretching vibration, 1690–1630 per cm) and amide-II vibration (N–H bending (40–60%) and C–N stretching vibrations (18–40%), 1655–1590 per cm) were the most dominant vibrational modes of amides [27]. The peak of C=O in 1650 per cm disappeared after chelation. It indicated that the oxygen atom of C=O may be chelated with Zn(II). The peaks of RNH2 and NH in 3450 and 3300 per cm shifted to high WN respectively, blue-shift. According to the structure of WP-10, this may be because the nitrogen atom of RNH2 and NH was chelated with Zn(II) leading to the increase of electronegativity around the nitrogen atom. The peaks of C–C(O) in 1200 per cm, represented aromatic acyl halide, shifted to low WN, known as red-shift. It is owing to the presence of the phenolic hydroxyl group of the tyrosine forming the hydrogen bond. This result confirmed that the absorption value of UV decreased at 220 nm and 280 nm after chelation. The FTIR spectra of peptide-zinc(II) complex in walnut showed similar changes [16].

Infrared spectrogram of the decapeptide and zinc-chelating peptide. The spectra were recorded between 4000 and 400 per cm at a resolution of 4 per cm with an average of 16 scans.
3.5. Molecular Dynamic Simulation of Zinc-Chelating Peptide
Molecular docking predicts the interactions between ligands and receptors by calculating interaction such as interaction energies, binding sites, and other information. As shown in Figure 5, the structure of WP-10 after energy minimization was used to simulate the docking of Zn(II). The docking results indicated that there were four possible binding sites of Zn(II) in WP-10. GLU1, PRO2, and ASN3 (GLU1PRO2ASN3) formed one of sites. GLU1 and ASN3 (GLU1ASN3) formed one site. ASN3 (ASN3) formed another site, while GLY4 and LEU6 (GLY4LEU6) form the last one. However, the chelation strength is different at different sites. The conclusion was also demonstrated in Figure 1.

Molecular docking simulation of peptide chelated with zinc. (A) was the structure of peptide after energy minimization. (B–E) were the chelation sites.
In Figure 6, the functional groups that play a major role in chelation was shown. As to WP-10, carbonyl group was the main functional groups interacting with Zn(II). The hydroxy group of GLU1 and the amidogen of ASN3 and LEU6 also could affect the chelation. According to these results, the structure of WP-10 could be changed and combined by site-specific mutagenesis for further application.

The chemical structure diagram of peptide. The structures marked with red color were represented the amino acids that participated in the chelation (marked with green name). The structures marked with black color were represented the amino acids that do not participated in the chelation (marked with purple name). The groups marked with blue color were represented the functional groups that participated in the chelation. The groups that may affect the chelation results were marked with pink color (repulsive force).
4. CONCLUSION
The WP-10 has an outstanding Zn(II) binding affinity. There are four sites in WP-10 that could bind with Zn(II) in Zn(II) rich environments. The maximal ratio of WP-10 chelated with Zn(II) was 1:1. GLU1, PRO2, ASN3, GLY4, and LEU6 were the main amino acids to form the chelation sites. The main groups involved in chelation were carbonyl groups. Some of the hydroxyl and amidogen groups also contribute to the chelation. The results from this study indicate that it is feasible to produce natural metal chelating using WP-10 peptides from walnut protein.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest.
AUTHORS’ CONTRIBUTION
WF contributed in writing original draft preparation. LJ and HRE contributed in writing review and editing. MD and CW contributed in supervision. ZW and ZM contributed in funding acquisition.
ACKNOWLEDGMENTS
This study was financially supported by the
Footnotes
REFERENCES
Cite this article
TY - JOUR AU - Weiwei Fan AU - Zhenyu Wang AU - Zhishen Mu AU - Ming Du AU - Lianzhou Jiang AU - Hesham R. EI-Seedi AU - Cong Wang PY - 2020 DA - 2020/08/03 TI - Characterizations of a Food Decapeptide Chelating with Zn(II) JO - eFood SP - 326 EP - 331 VL - 1 IS - 4 SN - 2666-3066 UR - https://doi.org/10.2991/efood.k.200727.001 DO - 10.2991/efood.k.200727.001 ID - Fan2020 ER -
